+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14066 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Rab GEF complex Mon1-Ccz1 | |||||||||
 Map data Map data | Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665; CtCcz1 aa1-796delta361-460). Map sharpened by Phenix local anisotropic filtering. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein targeting to vacuole / multivesicular body membrane / vacuolar membrane / vesicle-mediated transport /  autophagy autophagySimilarity search - Function | |||||||||
| Biological species |   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | |||||||||
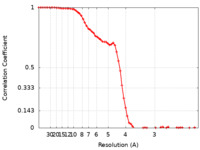
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.85 Å cryo EM / Resolution: 3.85 Å | |||||||||
 Authors Authors | Klink BU / Herrmann E / Antoni C / Langemeyer L / Kiontke S / Gatsogiannis C / Ungermann C / Raunser S / Kuemmel D | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation. Authors: Björn U Klink / Eric Herrmann / Claudia Antoni / Lars Langemeyer / Stephan Kiontke / Christos Gatsogiannis / Christian Ungermann / Stefan Raunser / Daniel Kümmel /  Abstract: Activation of the GTPase Rab7/Ypt7 by its cognate guanine nucleotide exchange factor (GEF) Mon1-Ccz1 marks organelles such as endosomes and autophagosomes for fusion with lysosomes/vacuoles and ...Activation of the GTPase Rab7/Ypt7 by its cognate guanine nucleotide exchange factor (GEF) Mon1-Ccz1 marks organelles such as endosomes and autophagosomes for fusion with lysosomes/vacuoles and degradation of their content. Here, we present a high-resolution cryogenic electron microscopy structure of the Mon1-Ccz1 complex that reveals its architecture in atomic detail. Mon1 and Ccz1 are arranged side by side in a pseudo-twofold symmetrical heterodimer. The three Longin domains of each Mon1 and Ccz1 are triangularly arranged, providing a strong scaffold for the catalytic center of the GEF. At the opposite side of the Ypt7-binding site, a positively charged and relatively flat patch stretches the Longin domains 2/3 of Mon1 and functions as a phosphatidylinositol phosphate-binding site, explaining how the GEF is targeted to membranes. Our work provides molecular insight into the mechanisms of endosomal Rab activation and serves as a blueprint for understanding the function of members of the Tri Longin domain Rab-GEF family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14066.map.gz emd_14066.map.gz | 20.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14066-v30.xml emd-14066-v30.xml emd-14066.xml emd-14066.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14066_fsc.xml emd_14066_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_14066.png emd_14066.png | 77.3 KB | ||
| Others |  emd_14066_additional_1.map.gz emd_14066_additional_1.map.gz emd_14066_additional_2.map.gz emd_14066_additional_2.map.gz emd_14066_half_map_1.map.gz emd_14066_half_map_1.map.gz emd_14066_half_map_2.map.gz emd_14066_half_map_2.map.gz | 19.2 MB 20.8 MB 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14066 http://ftp.pdbj.org/pub/emdb/structures/EMD-14066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14066 | HTTPS FTP |
-Related structure data
| Related structure data |  7qlaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14066.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14066.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
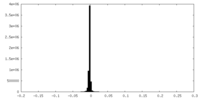
| Annotation | Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665; CtCcz1 aa1-796delta361-460). Map sharpened by Phenix local anisotropic filtering. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
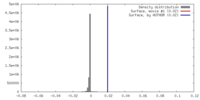
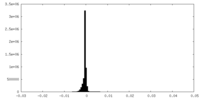
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665;...
| File | emd_14066_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
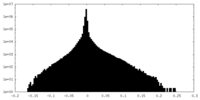
| Annotation | Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665; CtCcz1 aa1-796delta361-460). Map created by DeepEMhancer with high resolution training model. | ||||||||||||
| Projections & Slices |
| ||||||||||||
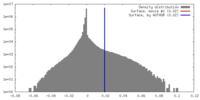
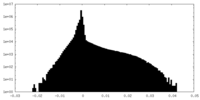
| Density Histograms |
-Additional map: Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665;...
| File | emd_14066_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
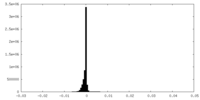
| Annotation | Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665; CtCcz1 aa1-796delta361-460). Map created by Relion Postprocessing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
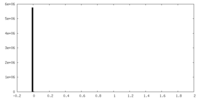
| Density Histograms |
-Half map: Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665;...
| File | emd_14066_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
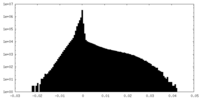
| Annotation | Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665; CtCcz1 aa1-796delta361-460). Unfiltered half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
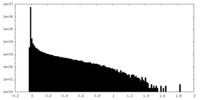
| Density Histograms |
-Half map: Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665;...
| File | emd_14066_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the Mon1-Ccz1 complex (CtMon1 aa141-665; CtCcz1 aa1-796delta361-460). Unfiltered half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rab GEF complex Mon1-Ccz1
| Entire | Name: Rab GEF complex Mon1-Ccz1 |
|---|---|
| Components |
|
-Supramolecule #1: Rab GEF complex Mon1-Ccz1
| Supramolecule | Name: Rab GEF complex Mon1-Ccz1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: CtMon1 aa141-665; CtCcz1 aa1-796delta361-460 |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: BL21 Escherichia coli (E. coli) / Recombinant strain: BL21 |
-Macromolecule #1: Vacuolar fusion protein MON1
| Macromolecule | Name: Vacuolar fusion protein MON1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 57.571832 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: TGTAGDLASL LAGDGLGRKS KAWRVLRAQQ QACGEGSGEE GEITEIEGLG LGEGMEGFER ELDNIPDTLP DDERLALWKG KLKHYLILS SAGKPIWSRH GDLSLVNSTM GVVQTIISFY EGARNPLLGF TAGKVRFVIL IKGPLYFVAI SRLRESDAQL R AQLEALYM ...String: TGTAGDLASL LAGDGLGRKS KAWRVLRAQQ QACGEGSGEE GEITEIEGLG LGEGMEGFER ELDNIPDTLP DDERLALWKG KLKHYLILS SAGKPIWSRH GDLSLVNSTM GVVQTIISFY EGARNPLLGF TAGKVRFVIL IKGPLYFVAI SRLRESDAQL R AQLEALYM QILSTLTLPI LTNIFAHRPS TDLRGPLQGT ESLLASLADS FTKGSPSTLL SALECLRLRK SQRQAITNIF LK SRCEELL YGLLVAGGKL VSVIRPRKHS LHPSDLQLIF NMLFESGGIK GNGGENWIPL CLPAFNNTGY LYMYVSFLDD KAP DDQNQP PESSNLDASN KNSSNTPDDD LTALILISPS REAFYALQSM RTRLVSQLLS TGYLSLIRST ALSGRPSITS ILPK TPLLH FLYKSRPNVQ WCMSSLSSLT PPGATATETL LARRKLMSVY EELHAALHAR HAHLRVVYST ADEKEGEGLA CLGWS TPAF EVYCVAPGCV GRAGMAREVN RVVQWARREE ERLFILGGGV F |
-Macromolecule #2: Ccz1
| Macromolecule | Name: Ccz1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Chaetomium thermophilum (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 75.780461 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTTPVSPSPS GIIPAQLGFL AIYNPALGTT DETLEDQIVY YATASTLSQA RRRHRRPRRR DRQRAQSVVK DSRPNAAGAT GDSEAVAED KDPVSKEERH ERLRQIGLAQ GMVEFAKSFS DGEPVDTIDT EKARVILVEV EEGWWILASI DLTRLPLPQI K TPTSSSAP ...String: MTTPVSPSPS GIIPAQLGFL AIYNPALGTT DETLEDQIVY YATASTLSQA RRRHRRPRRR DRQRAQSVVK DSRPNAAGAT GDSEAVAED KDPVSKEERH ERLRQIGLAQ GMVEFAKSFS DGEPVDTIDT EKARVILVEV EEGWWILASI DLTRLPLPQI K TPTSSSAP PPAPNLNPLP PEPAYEYSSR EVKPPSLLRA DLLRAYDLFL LHHGSSLSSL LASQGRAQLV ASLTRFWDHF LA TWNVLLH GNPACDVFGG IKLAASGELG IGVGEEERGS GEREVLEGLV ERVEGLVDVV VGRYGGPPSE KGPEEEQWLG LGG EVGEED GAVFLGVGAL DRKSLRGVVQ WMEEVYVWGE NAFGKPRRDL STGHFLLGLS ECSEEELTSS QANPKAIFVE LKPS YQHPS RKIPPEDPQP LGKVGPELPR DHTARLRPVI YVSQPFIYIL LFSEITPSPS TWPTLAESLH AQLSPLQKPL LHSTS YRPE RPVVETTSSS GTTTQHQIFD LVYDTETLTL QSTIPNIPDP FPYSATTPTG HSTGQQHHQQ SIWTRVEALQ THAQIL AIL SSGRAIPTDP SSFTHLPWEE GERTCKTARG WWIVWTRVVE HSPPDAVSLH HARDDDDNDD DASCSVLGHL RSVSSSH AA GSTSSSSGSG FGLGAIPGLG GLGGWAADGA TRLAQGIGID TRRYVEGLLT SLGR |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 0.001 mm Bright-field microscopy / Cs: 0.001 mm |
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 11916 / Average exposure time: 15.0 sec. / Average electron dose: 73.0 e/Å2 Details: Images were collected in movie-mode with 4 frames per second |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7qla: |
 Movie
Movie Controller
Controller














 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)