[English] 日本語
 Yorodumi
Yorodumi- EMDB-12762: CryoEM reveals BIN1 (isoform 8) does not bind to single actin fil... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12762 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reveals BIN1 (isoform 8) does not bind to single actin filaments in vitro | |||||||||
 Map data Map data | rabbit actin, polymerized, helical reconstruction with cryosparc 3.1 Helical twist -166.594 Helical rise 27.657 Angstrom | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity /  tropomyosin binding / tropomyosin binding /  myosin heavy chain binding / mesenchyme migration / myosin heavy chain binding / mesenchyme migration /  troponin I binding / actin filament bundle / filamentous actin / actin filament bundle assembly / skeletal muscle thin filament assembly / striated muscle thin filament ...cytoskeletal motor activator activity / troponin I binding / actin filament bundle / filamentous actin / actin filament bundle assembly / skeletal muscle thin filament assembly / striated muscle thin filament ...cytoskeletal motor activator activity /  tropomyosin binding / tropomyosin binding /  myosin heavy chain binding / mesenchyme migration / myosin heavy chain binding / mesenchyme migration /  troponin I binding / actin filament bundle / filamentous actin / actin filament bundle assembly / skeletal muscle thin filament assembly / striated muscle thin filament / skeletal muscle myofibril / actin monomer binding / skeletal muscle fiber development / troponin I binding / actin filament bundle / filamentous actin / actin filament bundle assembly / skeletal muscle thin filament assembly / striated muscle thin filament / skeletal muscle myofibril / actin monomer binding / skeletal muscle fiber development /  stress fiber / stress fiber /  titin binding / actin filament polymerization / titin binding / actin filament polymerization /  filopodium / filopodium /  actin filament / actin filament /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding /  lamellipodium / lamellipodium /  cell body / cell body /  hydrolase activity / protein domain specific binding / hydrolase activity / protein domain specific binding /  calcium ion binding / positive regulation of gene expression / magnesium ion binding / calcium ion binding / positive regulation of gene expression / magnesium ion binding /  ATP binding / identical protein binding / ATP binding / identical protein binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) | |||||||||
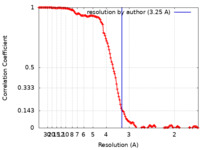
| Method | helical reconstruction /  cryo EM / Resolution: 3.25 Å cryo EM / Resolution: 3.25 Å | |||||||||
 Authors Authors | Wang Z / Mim C | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: MicroPubl Biol / Year: 2021 Journal: MicroPubl Biol / Year: 2021Title: CryoEM reveals BIN1 (isoform 8) does not bind to single actin filaments . Authors: Zuoneng Wang / Carsten Mim /  Abstract: Cells change their appearance by a concerted action of the cytoskeleton and the plasma membrane. The machinery that bends the membrane includes Bin/Amphiphysin/Rvs (BAR) domain proteins. Recently BAR ...Cells change their appearance by a concerted action of the cytoskeleton and the plasma membrane. The machinery that bends the membrane includes Bin/Amphiphysin/Rvs (BAR) domain proteins. Recently BAR domain proteins garnered attention as actin regulators, either by recruiting actin regulating proteins or through binding to actin directly. BIN1 (an important protein in Alzheimer's Disease, heart disease and cancer) is one of the few BAR proteins that bind to actin directly. Here, we imaged a complex of BIN1 and actin with cryoEM. Our results reveal that BIN1 cannot be found on single actin filaments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12762.map.gz emd_12762.map.gz | 24.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12762-v30.xml emd-12762-v30.xml emd-12762.xml emd-12762.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12762_fsc.xml emd_12762_fsc.xml | 13.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12762.png emd_12762.png | 62.7 KB | ||
| Others |  emd_12762_half_map_1.map.gz emd_12762_half_map_1.map.gz emd_12762_half_map_2.map.gz emd_12762_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12762 http://ftp.pdbj.org/pub/emdb/structures/EMD-12762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12762 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12762.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12762.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rabbit actin, polymerized, helical reconstruction with cryosparc 3.1 Helical twist -166.594 Helical rise 27.657 Angstrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_12762_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
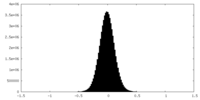
| Density Histograms |
-Half map: #2
| File | emd_12762_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
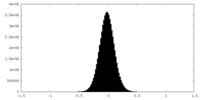
| Density Histograms |
- Sample components
Sample components
-Entire : Actin
| Entire | Name: Actin |
|---|---|
| Components |
|
-Supramolecule #1: Actin
| Supramolecule | Name: Actin / type: complex / ID: 1 / Parent: 0 / Details: Actin and BIN1 complex |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Experimental: 43 kDa/nm |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
Details: DTT was added fresh | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: 20mA, PelCo EasiGlo | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | 30 micromolar BIN1 + 15 micromolar actin |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 4621 / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X