[English] 日本語
 Yorodumi
Yorodumi- EMDB-12546: Structure of the darobactin-bound E. coli BAM complex (BamABCDE) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12546 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationBam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / cell outer membrane / protein-macromolecule adaptor activity /  cell adhesion / response to antibiotic / cell adhesion / response to antibiotic /  cell surface / cell surface /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli K-12 (bacteria) / Escherichia coli K-12 (bacteria) /   Escherichia coli (strain K12) (bacteria) / synthetic construct (others) Escherichia coli (strain K12) (bacteria) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.03 Å cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Jakob RP / Kaur H / Marzinek JK / Green R / Imai Y / Bolla J / Robinson C / Bond PJ / Lewis K / Maier T / Hiller S | |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Authors: Hundeep Kaur / Roman P Jakob / Jan K Marzinek / Robert Green / Yu Imai / Jani Reddy Bolla / Elia Agustoni / Carol V Robinson / Peter J Bond / Kim Lewis / Timm Maier / Sebastian Hiller /     Abstract: Antibiotics that target Gram-negative bacteria in new ways are needed to resolve the antimicrobial resistance crisis. Gram-negative bacteria are protected by an additional outer membrane, rendering ...Antibiotics that target Gram-negative bacteria in new ways are needed to resolve the antimicrobial resistance crisis. Gram-negative bacteria are protected by an additional outer membrane, rendering proteins on the cell surface attractive drug targets. The natural compound darobactin targets the bacterial insertase BamA-the central unit of the essential BAM complex, which facilitates the folding and insertion of outer membrane proteins. BamA lacks a typical catalytic centre, and it is not obvious how a small molecule such as darobactin might inhibit its function. Here we resolve the mode of action of darobactin at the atomic level using a combination of cryo-electron microscopy, X-ray crystallography, native mass spectrometry, in vivo experiments and molecular dynamics simulations. Two cyclizations pre-organize the darobactin peptide in a rigid β-strand conformation. This creates a mimic of the recognition signal of native substrates with a superior ability to bind to the lateral gate of BamA. Upon binding, darobactin replaces a lipid molecule from the lateral gate to use the membrane environment as an extended binding pocket. Because the interaction between darobactin and BamA is largely mediated by backbone contacts, it is particularly robust against potential resistance mutations. Our results identify the lateral gate as a functional hotspot in BamA and will allow the rational design of antibiotics that target this bacterial Achilles heel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12546.map.gz emd_12546.map.gz | 141.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12546-v30.xml emd-12546-v30.xml emd-12546.xml emd-12546.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12546.png emd_12546.png | 150.4 KB | ||
| Others |  emd_12546_additional_1.map.gz emd_12546_additional_1.map.gz emd_12546_half_map_1.map.gz emd_12546_half_map_1.map.gz emd_12546_half_map_2.map.gz emd_12546_half_map_2.map.gz | 267 MB 262.4 MB 262.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12546 http://ftp.pdbj.org/pub/emdb/structures/EMD-12546 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12546 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12546 | HTTPS FTP |
-Related structure data
| Related structure data |  7nriMC 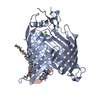 7nreC  7nrfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12546.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12546.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: sharpened map (B-Factor of 51.2)
| File | emd_12546_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened_map (B-Factor of 51.2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_12546_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_12546_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BamABCDE complex
| Entire | Name: BamABCDE complex |
|---|---|
| Components |
|
-Supramolecule #1: BamABCDE complex
| Supramolecule | Name: BamABCDE complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#5 |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) / Recombinant strain: C41 / Recombinant plasmid: pQLink Escherichia coli BL21(DE3) (bacteria) / Recombinant strain: C41 / Recombinant plasmid: pQLink |
-Macromolecule #1: Outer membrane protein assembly factor BamA
| Macromolecule | Name: Outer membrane protein assembly factor BamA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 88.22743 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVRVLRD GDTLLVQVKE RPTIASITFS GNKSVKDDM LKQNLEASGV RVGESLDRTT IADIEKGLED FYYSVGKYSA SVKAVVTPLP RNRVDLKLVF QEGVSAEIQQ I NIVGNHAF ...String: AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFATG NFEDVRVLRD GDTLLVQVKE RPTIASITFS GNKSVKDDM LKQNLEASGV RVGESLDRTT IADIEKGLED FYYSVGKYSA SVKAVVTPLP RNRVDLKLVF QEGVSAEIQQ I NIVGNHAF TTDELISHFQ LRDEVPWWNV VGDRKYQKQK LAGDLETLRS YYLDRGYARF NIDSTQVSLT PDKKGIYVTV NI TEGDQYK LSGVEVSGNL AGHSAEIEQL TKIEPGELYN GTKVTKMEDD IKKLLGRYGY AYPRVQSMPE INDADKTVKL RVN VDAGNR FYVRKIRFEG NDTSKDAVLR REMRQMEGAW LGSDLVDQGK ERLNRLGFFE TVDTDTQRVP GSPDQVDVVY KVKE RNTGS FNFGIGYGTE SGVSFQAGVQ QDNWLGTGYA VGINGTKNDY QTYAELSVTN PYFTVDGVSL GGRLFYNDFQ ADDAD LSDY TNKSYGTDVT LGFPINEYNS LRAGLGYVHN SLSNMQPQVA MWRYLYSMGE HPSTSDQDNS FKTDDFTFNY GWTYNK LDR GYFPTDGSRV NLTGKVTIPG SDNEYYKVTL DTATYVPIDD DHKWVVLGRT RWGYGDGLGG KEMPFYENFY AGGSSTV RG FQSNTIGPKA VYFPHQASNY DPDYDYECAT QDGAKDLCKS DDAVGGNAMA VASLEFITPT PFISDKYANS VRTSFFWD M GTVWDTNWDS SQYSGYPDYS DPSNIRMSAG IALQWMSPLG PLVFSYAQPF KKYDGDKAEQ FQFNIGK |
-Macromolecule #2: Outer membrane protein assembly factor BamB
| Macromolecule | Name: Outer membrane protein assembly factor BamB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 40.924516 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: CSLFNSEEDV VKMSPLPTVE NQFTPTTAWS TSVGSGIGNF YSNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEP ALLSGGVTVS GGHVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD G AVKWTVNL ...String: CSLFNSEEDV VKMSPLPTVE NQFTPTTAWS TSVGSGIGNF YSNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEP ALLSGGVTVS GGHVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD G AVKWTVNL DMPSLSLRGE SAPTTAFGAA VVGGDNGRVS AVLMEQGQMI WQQRISQATG STEIDRLSDV DTTPVVVNGV VF ALAYNGN LTALDLRSGQ IMWKRELGSV NDFIVDGNRI YLVDQNDRVM ALTIDGGVTL WTQSDLLHRL LTSPVLYNGN LVV GDSEGY LHWINVEDGR FVAQQKVDSS GFQTEPVAAD GKLLIQAKDG TVYSITRWSH PQFEK |
-Macromolecule #3: Outer membrane protein assembly factor BamC
| Macromolecule | Name: Outer membrane protein assembly factor BamC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 34.40125 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIRP PAQPLALVSG ARTQFTGDTA SLLVENGRG NTLWPQVVSV LQAKNYTITQ RDDAGQTLTT DWVQWNRLDE DEQYRGRYQI SVKPQGYQQA VTVKLLNLEQ A GKPVADAA ...String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIRP PAQPLALVSG ARTQFTGDTA SLLVENGRG NTLWPQVVSV LQAKNYTITQ RDDAGQTLTT DWVQWNRLDE DEQYRGRYQI SVKPQGYQQA VTVKLLNLEQ A GKPVADAA SMQRYSTEMM NVISAGLDKS ATDAANAAQN RASTTMDVQS AADDTGLPML VVRGPFNVVW QRLPAALEKV GM KVTDSTR SQGNMAVTYK PLSDSDWQEL GASDPGLASG DYKLQVGDLD NRSSLQFIDP KGHTLTQSQN DALVAVFQAA FSK |
-Macromolecule #4: Outer membrane protein assembly factor BamD
| Macromolecule | Name: Outer membrane protein assembly factor BamD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 25.816818 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: CSGSKEEVPD NPPNEIYATA QQKLQDGNWR QAITQLEALD NRYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYV MYMRGLTNMA LDDSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE Y SVAEYYTE ...String: CSGSKEEVPD NPPNEIYATA QQKLQDGNWR QAITQLEALD NRYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYV MYMRGLTNMA LDDSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE Y SVAEYYTE RGAWVAVVNR VEGMLRDYPD TQATRDALPL MENAYRQMQM NAQAEKVAKI IAANSSNT |
-Macromolecule #5: Outer membrane protein assembly factor BamE
| Macromolecule | Name: Outer membrane protein assembly factor BamE / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain K12) (bacteria) / Strain: K12 Escherichia coli (strain K12) (bacteria) / Strain: K12 |
| Molecular weight | Theoretical: 11.462772 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: CSTLERVVYR PDINQGNYLT ANDVSKIRVG MTQQQVAYAL GTPLMSDPFG TNTWFYVFRQ QPGHEGVTQQ TLTLTFNSSG VLTNIDNKP ALSGNKLHHH HHH |
-Macromolecule #6: 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE
| Macromolecule | Name: 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE / type: protein_or_peptide / ID: 6 / Details: Darobactin / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 971.047 Da |
| Sequence | String: WN(UX8)SKSF |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK III / Details: 4microL of sample applied. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.2 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | Manually grid screening, operated by SerialEM |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 5527 / Average exposure time: 9.0 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 870023 |
|---|---|
| CTF correction | Software - Name: cryoSPARC |
| Startup model | Type of model: OTHER / Details: cryoSpARC ab intio reconstruction |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: cryoSPARC |
| Final angle assignment | Type: OTHER / Software - Name: cryoSPARC |
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.03 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 157474 |
 Movie
Movie Controller
Controller
















 Z
Z Y
Y X
X