+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11892 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of native royal jelly filaments | |||||||||
 Map data Map data | native royal jelly filament | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcaste determination, influence by environmental factors / defense response to fungus / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to Gram-positive bacterium / extracellular region Similarity search - Function | |||||||||
| Biological species |   Apis mellifera (honey bee) / Apis mellifera (honey bee) /   Honeybee (honey bee) Honeybee (honey bee) | |||||||||
| Method | helical reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Mattei S / Ban A / Picenoni A / Leibundgut M / Glockshuber R / Boehringer D | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure of native glycolipoprotein filaments in honeybee royal jelly. Authors: Simone Mattei / Arvid Ban / Armin Picenoni / Marc Leibundgut / Rudi Glockshuber / Daniel Boehringer /   Abstract: Royal jelly (RJ) is produced by honeybees (Apis mellifera) as nutrition during larval development. The high viscosity of RJ originates from high concentrations of long lipoprotein filaments that ...Royal jelly (RJ) is produced by honeybees (Apis mellifera) as nutrition during larval development. The high viscosity of RJ originates from high concentrations of long lipoprotein filaments that include the glycosylated major royal jelly protein 1 (MRJP1), the small protein apisimin and insect lipids. Using cryo-electron microscopy we reveal the architecture and the composition of RJ filaments, in which the MRJP1 forms the outer shell of the assembly, surrounding stacked apisimin tetramers harbouring tightly packed lipids in the centre. The structural data rationalize the pH-dependent disassembly of RJ filaments in the gut of the larvae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11892.map.gz emd_11892.map.gz | 163.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11892-v30.xml emd-11892-v30.xml emd-11892.xml emd-11892.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11892.png emd_11892.png | 264.8 KB | ||
| Masks |  emd_11892_msk_1.map emd_11892_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_11892_half_map_1.map.gz emd_11892_half_map_1.map.gz emd_11892_half_map_2.map.gz emd_11892_half_map_2.map.gz | 138.8 MB 138.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11892 http://ftp.pdbj.org/pub/emdb/structures/EMD-11892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11892 | HTTPS FTP |
-Related structure data
| Related structure data |  7asdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11892.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11892.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | native royal jelly filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.19467 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11892_msk_1.map emd_11892_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: native royal jelly filament
| File | emd_11892_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | native royal jelly filament | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: native royal jelly filament
| File | emd_11892_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | native royal jelly filament | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : native royal jelly filaments
| Entire | Name: native royal jelly filaments |
|---|---|
| Components |
|
-Supramolecule #1: native royal jelly filaments
| Supramolecule | Name: native royal jelly filaments / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Apis mellifera (honey bee) Apis mellifera (honey bee) |
-Macromolecule #1: Major royal jelly protein 1
| Macromolecule | Name: Major royal jelly protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Honeybee (honey bee) Honeybee (honey bee) |
| Molecular weight | Theoretical: 48.934898 KDa |
| Sequence | String: MTRLFMLVCL GIVCQGTTGN ILRGESLNKS LPILHEWKFF DYDFGSDERR QDAILSGEYD YKNNYPSDID QWHDKIFVTM LRYNGVPSS LNVISKKVGD GGPLLQPYPD WSFAKYDDCS GIVSASKLAI DKCDRLWVLD SGLVNNTQPM CSPKLLTFDL T TSQLLKQV ...String: MTRLFMLVCL GIVCQGTTGN ILRGESLNKS LPILHEWKFF DYDFGSDERR QDAILSGEYD YKNNYPSDID QWHDKIFVTM LRYNGVPSS LNVISKKVGD GGPLLQPYPD WSFAKYDDCS GIVSASKLAI DKCDRLWVLD SGLVNNTQPM CSPKLLTFDL T TSQLLKQV EIPHDVAVNA TTGKGRLSSL AVQSLDCNTN SDTMVYIADE KGEGLIVYHN SDDSFHRLTS NTFDYDPKFT KM TIDGESY TAQDGISGMA LSPMTNNLYY SPVASTSLYY VNTEQFRTSD YQQNDIHYEG VQNILDTQSS AKVVSKSGVL FFG LVGDSA LGCWNEHRTL ERHNIRTVAQ SDETLQMIAS MKIKEALPHV PIFDRYINRE YILVLSNKMQ KMVNNDFNFD DVNF RIMNA NVNELILNTR CENPDNDRTP FKISIHL |
-Macromolecule #2: Apisimin
| Macromolecule | Name: Apisimin / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Honeybee (honey bee) Honeybee (honey bee) |
| Molecular weight | Theoretical: 7.949325 KDa |
| Sequence | String: MSKIVAVVVL AAFCVAMLVS DVSAKTSISV KGESNVDVVS QINSLVSSIV SGANVSAVLL AQTLVNILQI LIDANVFA |
-Macromolecule #5: (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol
| Macromolecule | Name: (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol / type: ligand / ID: 5 / Number of copies: 16 / Formula: 94R |
|---|---|
| Molecular weight | Theoretical: 398.664 Da |
| Chemical component information | 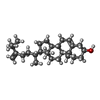 ChemComp-94R: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: SULFATE ION
| Macromolecule | Name: SULFATE ION / type: ligand / ID: 7 / Number of copies: 8 / Formula: SO4 |
|---|---|
| Molecular weight | Theoretical: 96.063 Da |
| Chemical component information |  ChemComp-SO4: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 119050 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 1.7 sec. / Average electron dose: 82.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: Gctf |
|---|---|
| Startup model | Type of model: OTHER / Details: featureless cylindrical density |
| Final angle assignment | Type: NOT APPLICABLE / Software - Name: RELION |
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 54.0 Å Applied symmetry - Helical parameters - Δ&Phi: 64 ° Applied symmetry - Helical parameters - Axial symmetry: D2 (2x2 fold dihedral  ) )Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 240483 |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X

























