[English] 日本語
 Yorodumi
Yorodumi- EMDB-10469: Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10469 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, peripheral stalk, rotational state 1 | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Euglena gracilis (euglena) Euglena gracilis (euglena) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Muhleip A / Amunts A | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Structure of a mitochondrial ATP synthase with bound native cardiolipin. Authors: Alexander Mühleip / Sarah E McComas / Alexey Amunts /  Abstract: The mitochondrial ATP synthase fuels eukaryotic cells with chemical energy. Here we report the cryo-EM structure of a divergent ATP synthase dimer from mitochondria of , a member of the phylum ...The mitochondrial ATP synthase fuels eukaryotic cells with chemical energy. Here we report the cryo-EM structure of a divergent ATP synthase dimer from mitochondria of , a member of the phylum Euglenozoa that also includes human parasites. It features 29 different subunits, 8 of which are newly identified. The membrane region was determined to 2.8 Å resolution, enabling the identification of 37 associated lipids, including 25 cardiolipins, which provides insight into protein-lipid interactions and their functional roles. The rotor-stator interface comprises four membrane-embedded horizontal helices, including a distinct subunit . The dimer interface is formed entirely by phylum-specific components, and a peripherally associated subcomplex contributes to the membrane curvature. The central and peripheral stalks directly interact with each other. Last, the ATPase inhibitory factor 1 (IF) binds in a mode that is different from human, but conserved in Trypanosomatids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
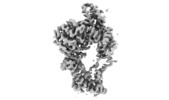
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10469.map.gz emd_10469.map.gz | 286.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10469-v30.xml emd-10469-v30.xml emd-10469.xml emd-10469.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10469.png emd_10469.png | 56.5 KB | ||
| Masks |  emd_10469_msk_1.map emd_10469_msk_1.map | 325 MB |  Mask map Mask map | |
| Others |  emd_10469_half_map_1.map.gz emd_10469_half_map_1.map.gz emd_10469_half_map_2.map.gz emd_10469_half_map_2.map.gz | 258.5 MB 258.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10469 http://ftp.pdbj.org/pub/emdb/structures/EMD-10469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10469 | HTTPS FTP |
-Related structure data
| Related structure data |  6tdwMC  6tduC  6tdvC  6tdxC  6tdyC  6tdzC  6te0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10469.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10469.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
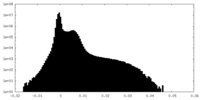
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10469_msk_1.map emd_10469_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10469_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10469_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Euglena gracilis mitochondrial ATP synthase dimer, perpipheral st...
| Entire | Name: Euglena gracilis mitochondrial ATP synthase dimer, perpipheral stalk region in rotational state 1 |
|---|---|
| Components |
|
-Supramolecule #1: Euglena gracilis mitochondrial ATP synthase dimer, perpipheral st...
| Supramolecule | Name: Euglena gracilis mitochondrial ATP synthase dimer, perpipheral stalk region in rotational state 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 2 MDa |
-Macromolecule #1: ATPTB1
| Macromolecule | Name: ATPTB1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 56.010633 KDa |
| Sequence | String: MTAVVKNLAK LPAATSQILT NVSKLQTFHG LLEERRDKYA PLAYHTYDNL KQKTTWHPIA HAWVDEGLPV SKKEYNEYCW LKKDMQRLL PLASPFVFGI YGILPLAVWL SNDGYLPSAF SSKKDIVSKK LEWYSSYGDD LRQQVGPMLQ HRLKRHLRGT L NNEHRLML ...String: MTAVVKNLAK LPAATSQILT NVSKLQTFHG LLEERRDKYA PLAYHTYDNL KQKTTWHPIA HAWVDEGLPV SKKEYNEYCW LKKDMQRLL PLASPFVFGI YGILPLAVWL SNDGYLPSAF SSKKDIVSKK LEWYSSYGDD LRQQVGPMLQ HRLKRHLRGT L NNEHRLML DEVTESYKEI FYSHYTGQLR DVRKCAHLRL YDGTSTVLLL TNKEPVELTS ELLQKWNAIK AAKLSPEEEK KA RNEALIE AYKEQELHGG PHVKHMQGYG IPSDTPLLGE NAKGDQYTQP PESASIPLEQ LEWTGDTVFI PAEYRTEVED WGR ELTKLA NQFLLLPWRF VSNAWNQRRL VSWFEEILQE DALIAKEGGV QALSDDELKV ALLDRAVIRC DEELTRGDME ARYK EISWL MSLRNPFIVL AWQTGYYRST YSPEDDLPEA SILPKLNRTV LDVDVHNELA PDHPEKPLPR VHPALYPNSH LALAK EVAV LAK |
-Macromolecule #2: subunit d
| Macromolecule | Name: subunit d / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 53.928289 KDa |
| Sequence | String: MMRRACRIIR PSHVRGVSGV APTIYLRSKA ALPATSTTDV RPQLYALQRF AKAQLKTATE AERAAIEADI ARYQEYLDSD LEKLKQDVA EDTAKKQKLI PLLDRYPDVP IEKIPEHANV LLKKIDACLE ILSKDIGEVT DAEAHEMYFE TSKFQILHIY T GCVASFPE ...String: MMRRACRIIR PSHVRGVSGV APTIYLRSKA ALPATSTTDV RPQLYALQRF AKAQLKTATE AERAAIEADI ARYQEYLDSD LEKLKQDVA EDTAKKQKLI PLLDRYPDVP IEKIPEHANV LLKKIDACLE ILSKDIGEVT DAEAHEMYFE TSKFQILHIY T GCVASFPE GDVPPGAVEC LPGQVIRTKV NGEDVMLEID EVDPGYQVCW FKPDVPLPEN AEILWSYPYE PTAALPTGTT WE EGQANVL IPAEPTPEAA VWPPTPVTNV YAPMAEKLAL KSNPELKVLF KEALLQPAKL LPLDVDYQCS HDREVVEAKR DRY LTALVE AEQAPPLPFT PDVLQLQLEH NVLKGELIDR LRALEYTIVT EQLQARLHER RLRGDVIDEW EELDYHPLVR DDTY LAIDF GDPTFGRYIW KLFPHTDGDE ECMFKDTRLD VLPPQVNPLN AILAQHTAQT PVHRSLEKRL WTEVRATAVS E |
-Macromolecule #3: ATPTB3
| Macromolecule | Name: ATPTB3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 35.175836 KDa |
| Sequence | String: MAAKTIPFVA STALGERLFA GVQKVLAAAK APVSLVQTDA KFAPADAKYL LAEPLSAAAT GELANKYGLS SKVTSGAASQ FYPNSIYPS LNVEVVQNLY ALSNPAFSSL SATTTKAVTV KDESAIKLAE LQKETERNIS SFFRDEANKS VQATLRLACD K AIKTKADK ...String: MAAKTIPFVA STALGERLFA GVQKVLAAAK APVSLVQTDA KFAPADAKYL LAEPLSAAAT GELANKYGLS SKVTSGAASQ FYPNSIYPS LNVEVVQNLY ALSNPAFSSL SATTTKAVTV KDESAIKLAE LQKETERNIS SFFRDEANKS VQATLRLACD K AIKTKADK KTVMVVTKPH GDAFDDLLAQ VTKSESDGRA DELRNSSVSV EPTLVGNAWP KLVMFPEGVN VVVCGPNASG DQ VAQLFVG IAGGTGMVAQ QLVGDAVVFT SANAEDNENP TGALLAASNL LTALGHEAEA KKIAAAVAKA YTTDRILPKE LPG GKADLE AFIDAVAKHA SA |
-Macromolecule #4: ATPTB4
| Macromolecule | Name: ATPTB4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 18.83351 KDa |
| Sequence | String: MFRGFRPVLA ADAVKFQTLY NVLTGKQHLK DQVPVKDCNL TAIFGASWKA DLNKWFDSEY APKLPAAERD SAKKSLDLYL KRVDLTRYT REELTTYGIL ACGPGKVDAL TEKHLLETGK ARLEELTAGL GNKDEGVNAF RKEVEQEGKY ANWPAEKSKA L ADKVIAAS P |
-Macromolecule #5: oligomycin sensitivity conferrring protein (OSCP)
| Macromolecule | Name: oligomycin sensitivity conferrring protein (OSCP) / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 29.56452 KDa |
| Sequence | String: MHMRRAVSVF GRCRSLNGLR NYAVPSPKYI EIYQSDFSRN AYPLELLGGS HVDFAKLLYS FADQVENKKF EVYVEDFKKL DSIIAEKGP FWAEEKIFQS PTFQGLSEGF KFILGWIQSE GAIDRLENVR LAYKELVNEA RKETTATVIV AKEPSGNDLA E IRKQVEEL ...String: MHMRRAVSVF GRCRSLNGLR NYAVPSPKYI EIYQSDFSRN AYPLELLGGS HVDFAKLLYS FADQVENKKF EVYVEDFKKL DSIIAEKGP FWAEEKIFQS PTFQGLSEGF KFILGWIQSE GAIDRLENVR LAYKELVNEA RKETTATVIV AKEPSGNDLA E IRKQVEEL HKESPLKDYK LVLETKVDPS IGGGYILEVC NQVVNRSAAA AAAETAALAK ASAAQVDWTS LPAAPPRPSP SA PDTLIRL LGSVVDDLAD ADKVEQKYGA |
-Macromolecule #6: subunit b
| Macromolecule | Name: subunit b / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 12.673109 KDa |
| Sequence | String: MPSTSPADKD VPMSILHTHG LSYVNWCMSL APGLLVFEGF FRARYYRSRV PPSRTVLMNG LKMRMFSLAR QQAPKIVHKP VLSPIPEHL RLVKNVAQVQ IDMLKLLNAQ AAK |
-Macromolecule #7: subunit 8
| Macromolecule | Name: subunit 8 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 7.112312 KDa |
| Sequence | String: LIPVSLVDLI NINIIFYILL LYTLLLFFIP LFLASINYTY HYIYKYYNYN YNFINNNT |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 3 seconds blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 130000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 9045 / Average exposure time: 10.0 sec. / Average electron dose: 36.3 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 555269 |
|---|---|
| CTF correction | Software - Name: RELION |
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final 3D classification | Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 150744 |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X