[English] 日本語
 Yorodumi
Yorodumi- EMDB-0126: Cryo-EM structure of the consensus hub of the clathrin coat complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0126 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the consensus hub of the clathrin coat complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  clathrin / coat protein / clathrin / coat protein /  endocytosis / trafficking / endocytosis / trafficking /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationclathrin coat of trans-Golgi network vesicle / clathrin light chain binding /  clathrin complex / clathrin coat of coated pit / clathrin complex / clathrin coat of coated pit /  clathrin-coated vesicle / vesicle-mediated transport / clathrin-coated vesicle / vesicle-mediated transport /  intracellular protein transport / disordered domain specific binding / intracellular protein transport / disordered domain specific binding /  nucleolus / structural molecule activity ...clathrin coat of trans-Golgi network vesicle / clathrin light chain binding / nucleolus / structural molecule activity ...clathrin coat of trans-Golgi network vesicle / clathrin light chain binding /  clathrin complex / clathrin coat of coated pit / clathrin complex / clathrin coat of coated pit /  clathrin-coated vesicle / vesicle-mediated transport / clathrin-coated vesicle / vesicle-mediated transport /  intracellular protein transport / disordered domain specific binding / intracellular protein transport / disordered domain specific binding /  nucleolus / structural molecule activity / nucleolus / structural molecule activity /  nucleoplasm / nucleoplasm /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Sus scrofa (pig) Sus scrofa (pig) | |||||||||
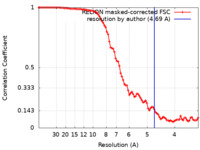
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.69 Å cryo EM / Resolution: 4.69 Å | |||||||||
 Authors Authors | Morris KL / Smith CJ | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly. Authors: Kyle L Morris / Joseph R Jones / Mary Halebian / Shenping Wu / Michael Baker / Jean-Paul Armache / Amaurys Avila Ibarra / Richard B Sessions / Alexander D Cameron / Yifan Cheng / Corinne J Smith /   Abstract: Clathrin forms diverse lattice and cage structures that change size and shape rapidly in response to the needs of eukaryotic cells during clathrin-mediated endocytosis and intracellular trafficking. ...Clathrin forms diverse lattice and cage structures that change size and shape rapidly in response to the needs of eukaryotic cells during clathrin-mediated endocytosis and intracellular trafficking. We present the cryo-EM structure and molecular model of assembled porcine clathrin, providing insights into interactions that stabilize key elements of the clathrin lattice, namely, between adjacent heavy chains, at the light chain-heavy chain interface and within the trimerization domain. Furthermore, we report cryo-EM maps for five different clathrin cage architectures. Fitting structural models to three of these maps shows that their assembly requires only a limited range of triskelion leg conformations, yet inherent flexibility is required to maintain contacts. Analysis of the protein-protein interfaces shows remarkable conservation of contact sites despite architectural variation. These data reveal a universal mode of clathrin assembly that allows variable cage architecture and adaptation of coated vesicle size and shape during clathrin-mediated vesicular trafficking or endocytosis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0126.map.gz emd_0126.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0126-v30.xml emd-0126-v30.xml emd-0126.xml emd-0126.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0126_fsc.xml emd_0126_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_0126.png emd_0126.png | 102.8 KB | ||
| Masks |  emd_0126_msk_1.map emd_0126_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0126.cif.gz emd-0126.cif.gz | 6.7 KB | ||
| Others |  emd_0126_additional_1.map.gz emd_0126_additional_1.map.gz emd_0126_additional_2.map.gz emd_0126_additional_2.map.gz emd_0126_half_map_1.map.gz emd_0126_half_map_1.map.gz emd_0126_half_map_2.map.gz emd_0126_half_map_2.map.gz | 60 MB 9 MB 14.8 MB 14.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0126 http://ftp.pdbj.org/pub/emdb/structures/EMD-0126 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0126 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0126 | HTTPS FTP |
-Related structure data
| Related structure data |  6sctMC  0114C  0115C  0116C  0118C  0120C  0121C  0122C  0123C  0124C  0125C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10296 (Title: Single particle cryo-EM dataset of the triskelion hub subparticle extraction from clathrin cages EMPIAR-10296 (Title: Single particle cryo-EM dataset of the triskelion hub subparticle extraction from clathrin cagesData size: 88.4 Data #1: Hub subparticles of the 28 mini coat [picked particles - multiframe - unprocessed] Data #2: Hub subparticles of the 32 sweet potato [picked particles - multiframe - unprocessed] Data #3: Hub subparticles of the 36 D6 barrel [picked particles - multiframe - unprocessed] Data #4: Hub subparticles of the 36 tennis ball [picked particles - multiframe - unprocessed] Data #5: Hub subparticles of the 37 big apple [picked particles - multiframe - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0126.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0126.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.705 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0126_msk_1.map emd_0126_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
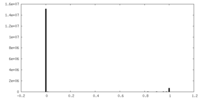
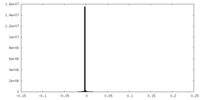
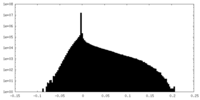
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Additional map used during model building at ad-hoc...
| File | emd_0126_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map used during model building at ad-hoc sharpened b-factor of -250. | ||||||||||||
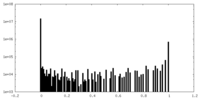
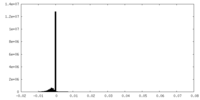
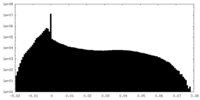
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Post processed map from resolution measurement.
| File | emd_0126_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post processed map from resolution measurement. | ||||||||||||
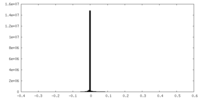
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Soft spherical masked (diameter 320A) applied to remove...
| File | emd_0126_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Soft spherical masked (diameter 320A) applied to remove artifactual density prior to resolution measurement. | ||||||||||||
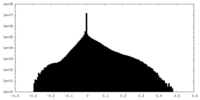
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Soft spherical masked (diameter 320A) applied to remove...
| File | emd_0126_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Soft spherical masked (diameter 320A) applied to remove artifactual density prior to resolution measurement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Assembly of clathrin heavy and light chain into coat complexes
| Entire | Name: Assembly of clathrin heavy and light chain into coat complexes |
|---|---|
| Components |
|
-Supramolecule #1: Assembly of clathrin heavy and light chain into coat complexes
| Supramolecule | Name: Assembly of clathrin heavy and light chain into coat complexes type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Sus scrofa (pig) / Organ: BRAIN Sus scrofa (pig) / Organ: BRAIN |
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: clathrin heavy chain
| Macromolecule | Name: clathrin heavy chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sus scrofa (pig) / Organ: BRAIN Sus scrofa (pig) / Organ: BRAIN |
| Sequence | String: MAQILPIRFQ EHLQLQNLGI NPANIGFSTL TMESDKFICI REKVGEQAQV VIIDMNDPSN PIRRPISAD SAIMNPASKV IALKAGKTLQ IFNIEMKSKM KAHTMTDDVT FWKWISLNTV A LVTDNAVY HWSMEGESQP VKMFDRHSSL AGCQIINYRT DAKQKWLLLT ...String: MAQILPIRFQ EHLQLQNLGI NPANIGFSTL TMESDKFICI REKVGEQAQV VIIDMNDPSN PIRRPISAD SAIMNPASKV IALKAGKTLQ IFNIEMKSKM KAHTMTDDVT FWKWISLNTV A LVTDNAVY HWSMEGESQP VKMFDRHSSL AGCQIINYRT DAKQKWLLLT GISAQQNRVV GA MQLYSVD RKVSQPIEGH AASFAQFKME GNAEESTLFC FAVRGQAGGK LHIIEVGTPP TGN QPFPKK AVDVFFPPEA QNDFPVAMQI SEKHDVVFLI TKYGYIHLYD LETGTCIYMN RISG ETIFV TAPHEATAGI IGVNRKGQVL SVCVEEENII PYITNVLQNP DLALRMAVRN NLAGA EELF ARKFNALFAQ GNYSEAAKVA ANAPKGILRT PDTIRRFQSV PAQPGQTSPL LQYFGI LLD QGQLNKYESL ELCRPVLQQG RKQLLEKWLK EDKLECSEEL GDLVKSVDPT LALSVYL RA NVPNKVIQCF AETGQVQKIV LYAKKVGYTP DWIFLLRNVM RISPDQGQQF AQMLVQDE E PLADITQIVD VFMEYNLIQQ CTAFLLDALK NNRPSEGPLQ TRLLEMNLMH APQVADAIL GNQMFTHYDR AHIAQLCEKA GLLQRALEHF TDLYDIKRAV VHTHLLNPEW LVNYFGSLSV EDSLECLRA MLSANIRQNL QICVQVASKY HEQLSTQSLI ELFESFKSFE GLFYFLGSIV N FSQDPDVH FKYIQAACKT GQIKEVERIC RESNCYDPER VKNFLKEAKL TDQLPLIIVC DR FDFVHDL VLYLYRNNLQ KYIEIYVQKV NPSRLPVVIG GLLDVDCSED VIKNLILVVR GQF STDELV AEVEKRNRLK LLLPWLEARI HEGCEEPATH NALAKIYIDS NNNPERFLRE NPYY DSRVV GKYCEKRDPH LACVAYERGQ CDLELINVCN ENSLFKSLSR YLVRRKDPEL WGSVL LESN PYRRPLIDQV VQTALSETQD PEEVSVTVKA FMTADLPNEL IELLEKIVLD NSVFSE HRN LQNLLILTAI KADRTRVMEY INRLDNYDAP DIANIAISNE LFEEAFAIFR KFDVNTS AV QVLIEHIGNL DRAYEFAERC NEPAVWSQLA KAQLQKGMVK EAIDSYIKAD DPSSYMEV V QAANTSGNWE ELVKYLQMAR KKARESYVET ELIFALAKTN RLAELEEFIN GPNNAHIQQ VGDRCYDEKM YDAAKLLYNN VSNFGRLAST LVHLGEYQAA VDGARKANST RTWKEVCFAC VDGKEFRLA QMCGLHIVVH ADELEELINY YQDRGYFEEL ITMLEAALGL ERAHMGMFTE L AILYSKFK PQKMREHLEL FWSRVNIPKV LRAAEQAHLW AELVFLYDKY EEYDNAIITM MN HPTDAWK EGQFKDIITK VANVELYYRA IQFYLEFKPL LLNDLLMVLS PRLDHTRAVN YFS KVKQLP LVKPYLRSVQ NHNNKSVNES LNNLFITEED YQALRTSIDA YDNFDNISLA QRLE KHELI EFRRIAAYLF KGNNRWKQSV ELCKKDSLYK DAMQYASESK DTELAEELLQ WFLQE EKRE CFGACLFTCY DLLRPDVVLE TAWRHNIMDF AMPYFIQVMK EYLTKVDKLD ASESLR KEE EQATETQPIV YGQPQLMLTA GPSVAVPPQA PFGYGYTAPP YGQPQPGFGY SM UniProtKB:  Clathrin heavy chain Clathrin heavy chain |
-Macromolecule #2: clathrin light chain b
| Macromolecule | Name: clathrin light chain b / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sus scrofa (pig) / Organ: BRAIN Sus scrofa (pig) / Organ: BRAIN |
| Sequence | String: MADDFGFFSS SESGAPEVAE EDPAAAFLAQ QESEIAGIEN DEGFGAPAGS QAALAQPGPA SGAGPEDMG TTVNGDVFQD ANGPADGYAA IAQADRLTQE PESIRKWREE QRKRLQELDA A SKVTEQEW REKAKKDLEE WNQRQSEQVE KNKINNRIAD KAFYQQPDAD ...String: MADDFGFFSS SESGAPEVAE EDPAAAFLAQ QESEIAGIEN DEGFGAPAGS QAALAQPGPA SGAGPEDMG TTVNGDVFQD ANGPADGYAA IAQADRLTQE PESIRKWREE QRKRLQELDA A SKVTEQEW REKAKKDLEE WNQRQSEQVE KNKINNRIAD KAFYQQPDAD IIGYVASEEA FV KESKEET PGTEWEKVAQ LCDFNPKSSK QCKDVSRLRS VLMSLKQTPL SR UniProtKB:  Nucleolar protein 16 Nucleolar protein 16 |
-Macromolecule #3: clathrin light chain a
| Macromolecule | Name: clathrin light chain a / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sus scrofa (pig) / Organ: BRAIN Sus scrofa (pig) / Organ: BRAIN |
| Sequence | String: MADLDPFGAP AGPSLGNGVA GEEDPAAAFL AQQESEIAGI ENDEAFAILD GGAPGPQPHG EPPGGPDAV DGVMNGEYYQ ESNGPTDSYA AISQVDRLQS EPESIRKWRE EQTERLEALD A NSRKQEAE WKEKAIKELE EWYARQDEQL QKTKANNRVA DEAFYKQPFA ...String: MADLDPFGAP AGPSLGNGVA GEEDPAAAFL AQQESEIAGI ENDEAFAILD GGAPGPQPHG EPPGGPDAV DGVMNGEYYQ ESNGPTDSYA AISQVDRLQS EPESIRKWRE EQTERLEALD A NSRKQEAE WKEKAIKELE EWYARQDEQL QKTKANNRVA DEAFYKQPFA DVIGYVAAEE AF VNDIEES SPGTEWERVA RLCDFNPKSS KQAKDVSRMR SVLISLKQAP LVH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.32 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.4 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: HOMEMADE PLUNGER Details: 3 uL applied to a grid at room temperature and humidity. 3 second hand blot and plunge.. | |||||||||||||||
| Details | Clathrin coat complexes, end point assembly exhibiting architectural heterogeneity |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 82111 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.4000000000000001 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.4000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average exposure time: 3.0 sec. / Average electron dose: 53.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6sct: |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X