[English] 日本語
 Yorodumi
Yorodumi- PDB-7yon: Complex structure of Neuropeptide Y Y2 receptor in complex with P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7yon | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  MEMBRANE PROTEIN / NPY / MEMBRANE PROTEIN / NPY /  GPCR / GPCR /  G-protein / G-protein /  Complex Complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptide YY receptor activity /  neuropeptide Y receptor activity / neuropeptide Y receptor activity /  neuropeptide Y receptor binding / intestinal epithelial cell differentiation / cardiac left ventricle morphogenesis / neuropeptide hormone activity / non-motile cilium / neuropeptide Y receptor binding / intestinal epithelial cell differentiation / cardiac left ventricle morphogenesis / neuropeptide hormone activity / non-motile cilium /  feeding behavior / outflow tract morphogenesis / calcium channel regulator activity ...peptide YY receptor activity / feeding behavior / outflow tract morphogenesis / calcium channel regulator activity ...peptide YY receptor activity /  neuropeptide Y receptor activity / neuropeptide Y receptor activity /  neuropeptide Y receptor binding / intestinal epithelial cell differentiation / cardiac left ventricle morphogenesis / neuropeptide hormone activity / non-motile cilium / neuropeptide Y receptor binding / intestinal epithelial cell differentiation / cardiac left ventricle morphogenesis / neuropeptide hormone activity / non-motile cilium /  feeding behavior / outflow tract morphogenesis / calcium channel regulator activity / Adenylate cyclase inhibitory pathway / neuropeptide signaling pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Peptide ligand-binding receptors / locomotory behavior / Regulation of insulin secretion / G protein-coupled receptor binding / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / feeding behavior / outflow tract morphogenesis / calcium channel regulator activity / Adenylate cyclase inhibitory pathway / neuropeptide signaling pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Peptide ligand-binding receptors / locomotory behavior / Regulation of insulin secretion / G protein-coupled receptor binding / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits /  cilium / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / cilium / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 /  hormone activity / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / response to peptide hormone / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / Inactivation, recovery and regulation of the phototransduction cascade / hormone activity / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / response to peptide hormone / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / Inactivation, recovery and regulation of the phototransduction cascade /  heterotrimeric G-protein complex / G alpha (12/13) signalling events / heterotrimeric G-protein complex / G alpha (12/13) signalling events /  extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  signaling receptor activity / signaling receptor activity /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway /  cell cortex / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / cell cortex / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling /  cell cycle / G protein-coupled receptor signaling pathway / lysosomal membrane / cell cycle / G protein-coupled receptor signaling pathway / lysosomal membrane /  cell division / cell division /  GTPase activity / GTPase activity /  centrosome / centrosome /  synapse / protein-containing complex binding / GTP binding / synapse / protein-containing complex binding / GTP binding /  nucleolus / magnesium ion binding / nucleolus / magnesium ion binding /  signal transduction / signal transduction /  extracellular space / extracellular exosome / extracellular region / extracellular space / extracellular exosome / extracellular region /  nucleoplasm / nucleoplasm /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)  Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.95 Å cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Kang, H. / Park, C. / Kim, J. / Choi, H.-J. | |||||||||
| Funding support |  Korea, Republic Of, 2items Korea, Republic Of, 2items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling. Authors: Hyunook Kang / Chaehee Park / Yeol Kyo Choi / Jungnam Bae / Sohee Kwon / Jinuk Kim / Chulwon Choi / Chaok Seok / Wonpil Im / Hee-Jung Choi /   Abstract: Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors ...Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors (YR and YR, respectively) activate G signaling, but their physiological responses to food intake are different. In addition, deletion of the two N-terminal amino acids of peptide YY (PYY(3-36)), the endogenous form found in circulation, can stimulate YR but not YR, suggesting that YR and YR may have distinct ligand-binding modes. Here, we report the cryo-electron microscopy structures of the PYY(3-36)‒YR‒G and NPY‒YR‒G complexes. Using cell-based assays, molecular dynamics simulations, and structural analysis, we revealed the molecular basis of the exclusive binding of PYY(3-36) to YR. Furthermore, we demonstrated that YR favors G protein signaling over β-arrestin signaling upon activation, whereas YR does not show a preference between these two pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7yon.cif.gz 7yon.cif.gz | 216.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7yon.ent.gz pdb7yon.ent.gz | 170.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7yon.json.gz 7yon.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yo/7yon https://data.pdbj.org/pub/pdb/validation_reports/yo/7yon ftp://data.pdbj.org/pub/pdb/validation_reports/yo/7yon ftp://data.pdbj.org/pub/pdb/validation_reports/yo/7yon | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 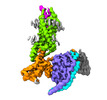 33984MC  7yooC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABG
| #1: Protein | Mass: 40415.031 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GNAI1 / Production host: Homo sapiens (human) / Gene: GNAI1 / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: P63096 Trichoplusia ni (cabbage looper) / References: UniProt: P63096 |
|---|---|
| #2: Protein | Mass: 37728.152 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: P62873 Trichoplusia ni (cabbage looper) / References: UniProt: P62873 |
| #3: Protein | Mass: 7861.143 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GNG2 / Production host: Homo sapiens (human) / Gene: GNG2 / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: P59768 Trichoplusia ni (cabbage looper) / References: UniProt: P59768 |
-Protein/peptide / Protein / Antibody / Non-polymers , 4 types, 5 molecules LRS

| #4: Protein/peptide | Mass: 4055.514 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) / References: UniProt: P10082 Homo sapiens (human) / References: UniProt: P10082 |
|---|---|
| #5: Protein | Mass: 43783.078 Da / Num. of mol.: 1 / Mutation: H149Y, S280C Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: NPY2R / Production host: Homo sapiens (human) / Gene: NPY2R / Production host:   Spodoptera frugiperda (fall armyworm) / References: UniProt: P49146 Spodoptera frugiperda (fall armyworm) / References: UniProt: P49146 |
| #6: Antibody | Mass: 27784.896 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| #7: Water | ChemComp-HOH /  Water Water |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| ||||||||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||||||||
| Specimen | Conc.: 10 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 278 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2000 nm / Nominal defocus min: 750 nm Bright-field microscopy / Nominal defocus max: 2000 nm / Nominal defocus min: 750 nm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 60.5 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
| EM imaging optics | Energyfilter name : GIF Bioquantum / Energyfilter slit width: 20 eV : GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 12262301 | ||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | ||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2.95 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 359459 / Symmetry type: POINT | ||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||
| Atomic model building |
| ||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj

































