[English] 日本語
 Yorodumi
Yorodumi- EMDB-27835: Cryo-EM structure of N-terminal arm (aa68-966) of BIRC6 (from loc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of N-terminal arm (aa68-966) of BIRC6 (from local refinement 3) | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationspongiotrophoblast layer development / labyrinthine layer development / ALK mutants bind TKIs / Flemming body /  microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / Signaling by ALK fusions and activated point mutants / microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / Signaling by ALK fusions and activated point mutants /  regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway ...spongiotrophoblast layer development / labyrinthine layer development / ALK mutants bind TKIs / Flemming body / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway ...spongiotrophoblast layer development / labyrinthine layer development / ALK mutants bind TKIs / Flemming body /  microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / Signaling by ALK fusions and activated point mutants / microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / Signaling by ALK fusions and activated point mutants /  regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / RING-type E3 ubiquitin transferase / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / RING-type E3 ubiquitin transferase /  trans-Golgi network / trans-Golgi network /  spindle pole / ubiquitin-protein transferase activity / regulation of cell population proliferation / midbody / cell population proliferation / protein ubiquitination / spindle pole / ubiquitin-protein transferase activity / regulation of cell population proliferation / midbody / cell population proliferation / protein ubiquitination /  endosome / endosome /  cell cycle / cell cycle /  cell division / cell division /  protein phosphorylation / protein phosphorylation /  centrosome / apoptotic process / positive regulation of cell population proliferation / negative regulation of apoptotic process / centrosome / apoptotic process / positive regulation of cell population proliferation / negative regulation of apoptotic process /  membrane / membrane /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
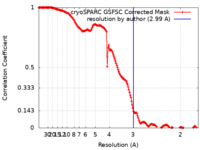
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.99 Å cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Hunkeler M / Fischer ES | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structures of BIRC6-client complexes provide a mechanism of SMAC-mediated release of caspases. Authors: Moritz Hunkeler / Cyrus Y Jin / Eric S Fischer /  Abstract: Tight regulation of apoptosis is essential for metazoan development and prevents diseases such as cancer and neurodegeneration. Caspase activation is central to apoptosis, and inhibitor of apoptosis ...Tight regulation of apoptosis is essential for metazoan development and prevents diseases such as cancer and neurodegeneration. Caspase activation is central to apoptosis, and inhibitor of apoptosis proteins (IAPs) are the principal actors that restrain caspase activity and are therefore attractive therapeutic targets. IAPs, in turn, are regulated by mitochondria-derived proapoptotic factors such as SMAC and HTRA2. Through a series of cryo-electron microscopy structures of full-length human baculoviral IAP repeat-containing protein 6 (BIRC6) bound to SMAC, caspases, and HTRA2, we provide a molecular understanding for BIRC6-mediated caspase inhibition and its release by SMAC. The architecture of BIRC6, together with near-irreversible binding of SMAC, elucidates how the IAP inhibitor SMAC can effectively control a processive ubiquitin ligase to respond to apoptotic stimuli. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27835.map.gz emd_27835.map.gz | 398.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27835-v30.xml emd-27835-v30.xml emd-27835.xml emd-27835.xml | 28.2 KB 28.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27835_fsc.xml emd_27835_fsc.xml | 19.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27835.png emd_27835.png | 26.2 KB | ||
| Masks |  emd_27835_msk_1.map emd_27835_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Others |  emd_27835_additional_1.map.gz emd_27835_additional_1.map.gz emd_27835_half_map_1.map.gz emd_27835_half_map_1.map.gz emd_27835_half_map_2.map.gz emd_27835_half_map_2.map.gz | 368.6 MB 391.6 MB 391.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27835 http://ftp.pdbj.org/pub/emdb/structures/EMD-27835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27835 | HTTPS FTP |
-Related structure data
| Related structure data |  8e2gMC  8e2dC  8e2eC  8e2fC  8e2hC  8e2iC  8e2jC  8e2kC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27835.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27835.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27835_msk_1.map emd_27835_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
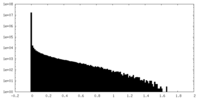
| Density Histograms |
-Additional map: main map post-processed using deepEMhancer
| File | emd_27835_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map post-processed using deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
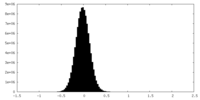
| Density Histograms |
-Half map: half-map 1
| File | emd_27835_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
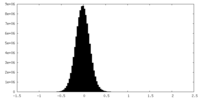
| Density Histograms |
-Half map: half-map 2
| File | emd_27835_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Baculoviral IAP repeat-containing protein 6
| Entire | Name: Baculoviral IAP repeat-containing protein 6 |
|---|---|
| Components |
|
-Supramolecule #1: Baculoviral IAP repeat-containing protein 6
| Supramolecule | Name: Baculoviral IAP repeat-containing protein 6 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.067 MDa |
-Macromolecule #1: Baculoviral IAP repeat-containing protein 6
| Macromolecule | Name: Baculoviral IAP repeat-containing protein 6 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  Ligases Ligases |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 534.158312 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKL AAANSSIDLI STSLYKKAGL TMVTGGGAAP PGTVTEPLPS VIVLSAGRKM AAAAAAASGP GCSSAAGAGA AGVSEWLVL RDGCMHCDAD GLHSLSYHPA LNAILAVTSR GTIKVIDGTS GATLQASALS AKPGGQVKCQ YISAVDKVIF V DDYAVGCR ...String: MDYKDDDDKL AAANSSIDLI STSLYKKAGL TMVTGGGAAP PGTVTEPLPS VIVLSAGRKM AAAAAAASGP GCSSAAGAGA AGVSEWLVL RDGCMHCDAD GLHSLSYHPA LNAILAVTSR GTIKVIDGTS GATLQASALS AKPGGQVKCQ YISAVDKVIF V DDYAVGCR KDLNGILLLD TALQTPVSKQ DDVVQLELPV TEAQQLLSAC LEKVDISSTE GYDLFITQLK DGLKNTSHET AA NHKVAKW ATVTFHLPHH VLKSIASAIV NELKKINQNV AALPVASSVM DRLSYLLPSA RPELGVGPGR SVDRSLMYSE ANR RETFTS WPHVGYRWAQ PDPMAQAGFY HQPASSGDDR AMCFTCSVCL VCWEPTDEPW SEHERHSPNC PFVKGEHTQN VPLS VTLAT SPAQFPCTDG TDRISCFGSG SCPHFLAAAT KRGKICIWDV SKLMKVHLKF EINAYDPAIV QQLILSGDPS SGVDS RRPT LAWLEDSSSC SDIPKLEGDS DDLLEDSDSE EHSRSDSVTG HTSQKEAMEV SLDITALSIL QQPEKLQWEI VANVLE DTV KDLEELGANP CLTNSKSEKT KEKHQEQHNI PFPCLLAGGL LTYKSPATSP ISSNSHRSLD GLSRTQGESI SEQGSTD NE SCTNSELNSP LVRRTLPVLL LYSIKESDEK AGKIFSQMNN IMSKSLHDDG FTVPQIIEME LDSQEQLLLQ DPPVTYIQ Q FADAAANLTS PDSEKWNSVF PKPGTLVQCL RLPKFAEEEN LCIDSITPCA DGIHLLVGLR TCPVESLSAI NQVEALNNL NKLNSALCNR RKGELESNLA VVNGANISVI QHESPADVQT PLIIQPEQRN VSGGYLVLYK MNYATRIVTL EEEPIKIQHI KDPQDTITS LILLPPDILD NREDDCEEPI EDMQLTSKNG FEREKTSDIS TLGHLVITTQ GGYVKILDLS NFEILAKVEP P KKEGTEEQ DTFVSVIYCS GTDRLCACTK GGELHFLQIG GTCDDIDEAD ILVDGSLSKG IEPSSEGSKP LSNPSSPGIS GV DLLVDQP FTLEILTSLV ELTRFETLTP RFSATVPPCW VEVQQEQQQR RHPQHLHQQH HGDAAQHTRT WKLQTDSNSW DEH VFELVL PKACMVGHVD FKFVLNSNIT NIPQIQVTLL KNKAPGLGKV NALNIEVEQN GKPSLVDLNE EMQHMDVEES QCLR LCPFL EDHKEDILCG PVWLASGLDL SGHAGMLTLT SPKLVKGMAG GKYRSFLIHV KAVNERGTEE ICNGGMRPVV RLPSL KHQS NKGYSLASLL AKVAAGKEKS SNVKNENTSG TRKSENLRGC DLLQEVSVTI RRFKKTSISK ERVQRCAMLQ FSEFHE KLV NTLCRKTDDG QITEHAQSLV LDTLCWLAGV HSNGPGSSKE GNENLLSKTR KFLSDIVRVC FFEAGRSIAH KCARFLA LC ISNGKCDPCQ PAFGPVLLKA LLDNMSFLPA ATTGGSVYWY FVLLNYVKDE DLAGCSTACA SLLTAVSRQL QDRLTPME A LLQTRYGLYS SPFDPVLFDL EMSGSSCKNV YNSSIGVQSD EIDLSDVLSG NGKVSSCTAA EGSFTSLTGL LEVEPLHFT CVSTSDGTRI ERDDAMSSFG VTPAVGGLSS GTVGEASTAL SSAAQVALQS LSHAMASAEQ QLQVLQEKQQ QLLKLQQQKA KLEAKLHQT TAAAAAAASA VGPVHNSVPS NPVAAPGFFI HPSDVIPPTP KTTPLFMTPP LTPPNEAVSV VINAELAQLF P GSVIDPPA VNLAAHNKNS NKSRMNPLGS GLALAISHAS HFLQPPPHQS IIIERMHSGA RRFVTLDFGR PILLTDVLIP TC GDLASLS IDIWTLGEEV DGRRLVVATD ISTHSLILHD LIPPPVCRFM KITVIGRYGS TNARAKIPLG FYYGHTYILP WES ELKLMH DPLKGEGESA NQPEIDQHLA MMVALQEDIQ CRYNLACHRL ETLLQSIDLP PLNSANNAQY FLRKPDKAVE EDSR VFSAY QDCIQLQLQL NLAHNAVQRL KVALGASRKM LSETSNPEDL IQTSSTEQLR TIIRYLLDTL LSLLHASNGH SVPAV LQST FHAQACEELF KHLCISGTPK IRLHTGLLLV QLCGGERWWG QFLSNVLQEL YNSEQLLIFP QDRVFMLLSC IGQRSL SNS GVLESLLNLL DNLLSPLQPQ LPMHRRTEGV LDIPMISWVV MLVSRLLDYV ATVEDEAAAA KKPLNGNQWS FINNNLH TQ SLNRSSKGSS SLDRLYSRKI RKQLVHHKQQ LNLLKAKQKA LVEQMEKEKI QSNKGSSYKL LVEQAKLKQA TSKHFKDL I RLRRTAEWSR SNLDTEVTTA KESPEIEPLP FTLAHERCIS VVQKLVLFLL SMDFTCHADL LLFVCKVLAR IANATRPTI HLCEIVNEPQ LERLLLLLVG TDFNRGDISW GGAWAQYSLT CMLQDILAGE LLAPVAAEAM EEGTVGDDVG ATAGDSDDSL QQSSVQLLE TIDEPLTHDI TGAPPLSSLE KDKEIDLELL QDLMEVDIDP LDIDLEKDPL AAKVFKPISS TWYDYWGADY G TYNYNPYI GGLGIPVAKP PANTEKNGSQ TVSVSVSQAL DARLEVGLEQ QAELMLKMMS TLEADSILQA LTNTSPTLSQ SP TGTDDSL LGGLQAANQT SQLIIQLSSV PMLNVCFNKL FSMLQVHHVQ LESLLQLWLT LSLNSSSTGN KENGADIFLY NAN RIPVIS LNQASITSFL TVLAWYPNTL LRTWCLVLHS LTLMTNMQLN SGSSSAIGTQ ESTAHLLVSD PNLIHVLVKF LSGT SPHGT NQHSPQVGPT ATQAMQEFLT RLQVHLSSTC PQIFSEFLLK LIHILSTERG AFQTGQGPLD AQVKLLEFTL EQNFE VVSV STISAVIESV TFLVHHYITC SDKVMSRSGS DSSVGARACF GGLFANLIRP GDAKAVCGEM TRDQLMFDLL KLVNIL VQL PLSGNREYSA RVSVTTNTTD SVSDEEKVSG GKDGNGSSTS VQGSPAYVAD LVLANQQIMS QILSALGLCN SSAMAMI IG ASGLHLTKHE NFHGGLDAIS VGDGLFTILT TLSKKASTVH MMLQPILTYM ACGYMGRQGS LATCQLSEPL LWFILRVL D TSDALKAFHD MGGVQLICNN MVTSTRAIVN TARSMVSTIM KFLDSGPNKA VDSTLKTRIL ASEPDNAEGI HNFAPLGTI TSSSPTAQPA EVLLQATPPH RRARSAAWSY IFLPEEAWCD LTIHLPAAVL LKEIHIQPHL ASLATCPSSV SVEVSADGVN MLPLSTPVV TSGLTYIKIQ LVKAEVASAV CLRLHRPRDA STLGLSQIKL LGLTAFGTTS SATVNNPFLP SEDQVSKTSI G WLRLLHHC LTHISDLEGM MASAAAPTAN LLQTCAALLM SPYCGMHSPN IEVVLVKIGL QSTRIGLKLI DILLRNCAAS GS DPTDLNS PLLFGRLNGL SSDSTIDILY QLGTTQDPGT KDRIQALLKW VSDSARVAAM KRSGRMNYMC PNSSTVEYGL LMP SPSHLH CVAAILWHSY ELLVEYDLPA LLDQELFELL FNWSMSLPCN MVLKKAVDSL LCSMCHVHPN YFSLLMGWMG ITPP PVQCH HRLSMTDDSK KQDLSSSLTD DSKNAQAPLA LTESHLATLA SSSQSPEAIK QLLDSGLPSL LVRSLASFCF SHISS SESI AQSIDISQDK LRRHHVPQQC NKMPITADLV APILRFLTEV GNSHIMKDWL GGSEVNPLWT ALLFLLCHSG STSGSH NLG AQQTSARSAS LSSAATTGLT TQQRTAIENA TVAFFLQCIS CHPNNQKLMA QVLCELFQTS PQRGNLPTSG NISGFIR RL FLQLMLEDEK VTMFLQSPCP LYKGRINATS HVIQHPMYGA GHKFRTLHLP VSTTLSDVLD RVSDTPSITA KLISEQKD D KEKKNHEEKE KVKAENGFQD NYSVVVASGL KSQSKRAVSA TPPRPPSRRG RTIPDKIGST SGAEAANKII TVPVFHLFH KLLAGQPLPA EMTLAQLLTL LYDRKLPQGY RSIDLTVKLG SRVITDPSLS KTDSYKRLHP EKDHGDLLAS CPEDEALTPG DECMDGILD ESLLETCPIQ SPLQVFAGMG GLALIAERLP MLYPEVIQQV SAPVVTSTTQ EKPKDSDQFE WVTIEQSGEL V YEAPETVA AEPPPIKSAV QTMSPIPAHS LAAFGLFLRL PGYAEVLLKE RKHAQCLLRL VLGVTDDGEG SHILQSPSAN VL PTLPFHV LRSLFSTTPL TTDDGVLLRR MALEIGALHL ILVCLSALSH HSPRVPNSSV NQTEPQVSSS HNPTSTEEQQ LYW AKGTGF GTGSTASGWD VEQALTKQRL EEEHVTCLLQ VLASYINPVS SAVNGEAQSS HETRGQNSNA LPSVLLELLS QSCL IPAMS SYLRNDSVLD MARHVPLYRA LLELLRAIAS CAAMVPLLLP LSTENGEEEE EQSECQTSVG TLLAKMKTCV DTYTN RLRS KRENVKTGVK PDASDQEPEG LTLLVPDIQK TAEIVYAATT SLRQANQEKK LGEYSKKAAM KPKPLSVLKS LEEKYV AVM KKLQFDTFEM VSEDEDGKLG FKVNYHYMSQ VKNANDANSA ARARRLAQEA VTLSTSLPLS SSSSVFVRCD EERLDIM KV LITGPADTPY ANGCFEFDVY FPQDYPSSPP LVNLETTGGH SVRFNPNLYN DGKVCLSILN TWHGRPEEKW NPQTSSFL Q VLVSVQSLIL VAEPYFNEPG YERSRGTPSG TQSSREYDGN IRQATVKWAM LEQIRNPSPC FKEVIHKHFY LKRVEIMAQ CEEWIADIQQ YSSDKRVGRT MSHHAAALKR HTAQLREELL KLPCPEGLDP DTDDAPEVCR ATTGAEETLM HDQVKPSSSK ELPSDFQL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283.15 K / Instrument: LEICA EM GP | ||||||||||||
| Details | added CHAPSO to 0.8 mM |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | Data collection in counting mode, using multi-shot scheme (9 holes per stage position, 3 movies per hole) |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 19647 / Average exposure time: 2.4 sec. / Average electron dose: 52.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8e2g: |
 Movie
Movie Controller
Controller















 Z
Z Y
Y X
X