[English] 日本語
 Yorodumi
Yorodumi- EMDB-26958: Local refinement of Rh trimer, glycophorin B and Band3-III transm... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
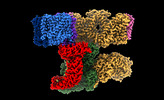
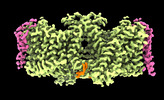
| Title | Local refinement of Rh trimer, glycophorin B and Band3-III transmembrane region, class 1a of erythrocyte ankyrin-1 complex | |||||||||
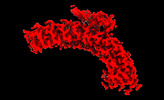
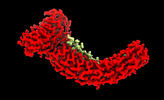
 Map data Map data | Main map used for model fitting. Density modified and cropped using phenix.resolve_cryo_em, resampled on fine grid using relion_image_handler. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmethylammonium transmembrane transport / Defective RHAG causes regulator type Rh-null hemolytic anemia (RHN) / Rhesus blood group biosynthesis / methylammonium transmembrane transporter activity / Rhesus glycoproteins mediate ammonium transport. / ammonium homeostasis / carbon dioxide transmembrane transport / carbon dioxide transmembrane transporter activity / pH elevation / ammonium transmembrane transport ...methylammonium transmembrane transport / Defective RHAG causes regulator type Rh-null hemolytic anemia (RHN) / Rhesus blood group biosynthesis / methylammonium transmembrane transporter activity / Rhesus glycoproteins mediate ammonium transport. / ammonium homeostasis / carbon dioxide transmembrane transport / carbon dioxide transmembrane transporter activity / pH elevation / ammonium transmembrane transport / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) /  leak channel activity / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis / leak channel activity / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis /  ankyrin-1 complex / plasma membrane phospholipid scrambling / monoatomic anion transmembrane transporter activity / chloride:bicarbonate antiporter activity / ammonium channel activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity / bicarbonate transport / monoatomic anion transport / inorganic cation transmembrane transport / chloride transmembrane transporter activity / chloride transport / erythrocyte development / negative regulation of glycolytic process through fructose-6-phosphate / ankyrin-1 complex / plasma membrane phospholipid scrambling / monoatomic anion transmembrane transporter activity / chloride:bicarbonate antiporter activity / ammonium channel activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity / bicarbonate transport / monoatomic anion transport / inorganic cation transmembrane transport / chloride transmembrane transporter activity / chloride transport / erythrocyte development / negative regulation of glycolytic process through fructose-6-phosphate /  ankyrin binding / ankyrin binding /  hemoglobin binding / cortical cytoskeleton / protein-membrane adaptor activity / chloride transmembrane transport / protein localization to plasma membrane / hemoglobin binding / cortical cytoskeleton / protein-membrane adaptor activity / chloride transmembrane transport / protein localization to plasma membrane /  regulation of intracellular pH / Cell surface interactions at the vascular wall / carbon dioxide transport / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / transmembrane transport / cytoplasmic side of plasma membrane / Z disc / multicellular organismal-level iron ion homeostasis / regulation of intracellular pH / Cell surface interactions at the vascular wall / carbon dioxide transport / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / transmembrane transport / cytoplasmic side of plasma membrane / Z disc / multicellular organismal-level iron ion homeostasis /  blood coagulation / virus receptor activity / basolateral plasma membrane / blood microparticle / protein homodimerization activity / extracellular exosome / blood coagulation / virus receptor activity / basolateral plasma membrane / blood microparticle / protein homodimerization activity / extracellular exosome /  nucleoplasm / nucleoplasm /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   human (human) human (human) | |||||||||
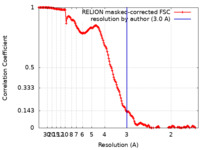
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Vallese F / Kim K / Yen LY / Johnston JD / Noble AJ / Cali T / Clarke OB | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Architecture of the human erythrocyte ankyrin-1 complex. Authors: Francesca Vallese / Kookjoo Kim / Laura Y Yen / Jake D Johnston / Alex J Noble / Tito Calì / Oliver Biggs Clarke /   Abstract: The stability and shape of the erythrocyte membrane is provided by the ankyrin-1 complex, but how it tethers the spectrin-actin cytoskeleton to the lipid bilayer and the nature of its association ...The stability and shape of the erythrocyte membrane is provided by the ankyrin-1 complex, but how it tethers the spectrin-actin cytoskeleton to the lipid bilayer and the nature of its association with the band 3 anion exchanger and the Rhesus glycoproteins remains unknown. Here we present structures of ankyrin-1 complexes purified from human erythrocytes. We reveal the architecture of a core complex of ankyrin-1, the Rhesus proteins RhAG and RhCE, the band 3 anion exchanger, protein 4.2, glycophorin A and glycophorin B. The distinct T-shaped conformation of membrane-bound ankyrin-1 facilitates recognition of RhCE and, unexpectedly, the water channel aquaporin-1. Together, our results uncover the molecular details of ankyrin-1 association with the erythrocyte membrane, and illustrate the mechanism of ankyrin-mediated membrane protein clustering. | |||||||||
| History |
|
- Structure visualization
Structure visualization
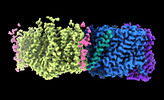
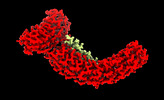
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26958.map.gz emd_26958.map.gz | 327.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26958-v30.xml emd-26958-v30.xml emd-26958.xml emd-26958.xml | 36.8 KB 36.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26958_fsc.xml emd_26958_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26958.png emd_26958.png | 96.1 KB | ||
| Others |  emd_26958_additional_1.map.gz emd_26958_additional_1.map.gz emd_26958_additional_2.map.gz emd_26958_additional_2.map.gz emd_26958_additional_3.map.gz emd_26958_additional_3.map.gz emd_26958_additional_4.map.gz emd_26958_additional_4.map.gz emd_26958_half_map_1.map.gz emd_26958_half_map_1.map.gz emd_26958_half_map_2.map.gz emd_26958_half_map_2.map.gz | 322.1 MB 864.7 KB 322.1 MB 328.1 MB 326.8 MB 326.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26958 http://ftp.pdbj.org/pub/emdb/structures/EMD-26958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26958 | HTTPS FTP |
-Related structure data
| Related structure data |  8crtMC  7uz3C  7uzeC  7uzqC  7uzsC  7uzuC  7uzvC  7v07C  7v0kC  7v0mC  7v0qC  7v0sC  7v0tC  7v0uC  7v0xC  7v0yC  7v19C  8crqC  8crrC  8cs9C  8cslC  8csvC  8cswC  8csxC  8csyC  8ct2C  8ct3C  8cteC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26958.map.gz / Format: CCP4 / Size: 352.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26958.map.gz / Format: CCP4 / Size: 352.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map used for model fitting. Density modified and cropped using phenix.resolve_cryo_em, resampled on fine grid using relion_image_handler. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.415 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Half map 2 (unmodified)
| File | emd_26958_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 (unmodified) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Mask used for FSC calculation
| File | emd_26958_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mask used for FSC calculation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half map 1 (unmodified)
| File | emd_26958_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 (unmodified) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor sharpened map.
| File | emd_26958_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 (cropped and resampled to match main map).
| File | emd_26958_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 (cropped and resampled to match main map). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 (cropped and resampled to match main map).
| File | emd_26958_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 (cropped and resampled to match main map). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Erythrocyte ankyrin-1 complex
| Entire | Name: Erythrocyte ankyrin-1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Erythrocyte ankyrin-1 complex
| Supramolecule | Name: Erythrocyte ankyrin-1 complex / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Purified by density gradient centrifugation and size exclusion chromatography from digitonin-solubilized erythrocyte membranes. |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Organ: Blood / Tissue: Erythrocytes / Location in cell: Plasma membrane Homo sapiens (human) / Organ: Blood / Tissue: Erythrocytes / Location in cell: Plasma membrane |
-Macromolecule #1: Blood group Rh(CE) polypeptide
| Macromolecule | Name: Blood group Rh(CE) polypeptide / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   human (human) human (human) |
| Molecular weight | Theoretical: 45.598918 KDa |
| Sequence | String: MSSKYPRSVR RCLPLWALTL EAALILLFYF FTHYDASLED QKGLVASYQV GQDLTVMAAL GLGFLTSNFR RHSWSSVAFN LFMLALGVQ WAILLDGFLS QFPPGKVVIT LFSIRLATMS AMSVLISAGA VLGKVNLAQL VVMVLVEVTA LGTLRMVISN I FNTDYHMN ...String: MSSKYPRSVR RCLPLWALTL EAALILLFYF FTHYDASLED QKGLVASYQV GQDLTVMAAL GLGFLTSNFR RHSWSSVAFN LFMLALGVQ WAILLDGFLS QFPPGKVVIT LFSIRLATMS AMSVLISAGA VLGKVNLAQL VVMVLVEVTA LGTLRMVISN I FNTDYHMN LRHFYVFAAY FGLTVAWCLP KPLPKGTEDN DQRATIPSLS AMLGALFLWM FWPSVNSPLL RSPIQRKNAM FN TYYALAV SVVTAISGSS LAHPQRKISM TYVHSAVLAG GVAVGTSCHL IPSPWLAMVL GLVAGLISIG GAKCLPVCCN RVL GIHHIS VMHSIFSLLG LLGEITYIVL LVLHTVWNGN GMIGFQVLLS IGELSLAIVI ALTSGLLTGL LLNLKIWKAP HVAK YFDDQ VFWKFPHLAV GF |
-Macromolecule #2: Ammonium transporter Rh type A
| Macromolecule | Name: Ammonium transporter Rh type A / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   human (human) human (human) |
| Molecular weight | Theoretical: 44.229629 KDa |
| Sequence | String: MRFTFPLMAI VLEIAMIVLF GLFVEYETDQ TVLEQLNITK PTDMGIFFEL YPLFQDVHVM IFVGFGFLMT FLKKYGFSSV GINLLVAAL GLQWGTIVQG ILQSQGQKFN IGIKNMINAD FSAATVLISF GAVLGKTSPT QMLIMTILEI VFFAHNEYLV S EIFKASDI ...String: MRFTFPLMAI VLEIAMIVLF GLFVEYETDQ TVLEQLNITK PTDMGIFFEL YPLFQDVHVM IFVGFGFLMT FLKKYGFSSV GINLLVAAL GLQWGTIVQG ILQSQGQKFN IGIKNMINAD FSAATVLISF GAVLGKTSPT QMLIMTILEI VFFAHNEYLV S EIFKASDI GASMTIHAFG AYFGLAVAGI LYRSGLRKGH ENEESAYYSD LFAMIGTLFL WMFWPSFNSA IAEPGDKQCR AI VNTYFSL AACVLTAFAF SSLVEHRGKL NMVHIQNATL AGGVAVGTCA DMAIHPFGSM IIGSIAGMVS VLGYKFLTPL FTT KLRIHD TCGVHNLHGL PGVVGGLAGI VAVAMGASNT SMAMQAAALG SSIGTAVVGG LMTGLILKLP LWGQPSDQNC YDDS VYWKV PKTR |
-Macromolecule #3: Glycophorin-B
| Macromolecule | Name: Glycophorin-B / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   human (human) human (human) |
| Molecular weight | Theoretical: 9.789674 KDa |
| Sequence | String: MYGKIIFVLL LSEIVSISAL STTEVAMHTS TSSSVTKSYI SSQTNGETGQ LVHRFTVPAP VVIILIILCV MAGIIGTILL ISYSIRRLI KA |
-Macromolecule #4: Glycophorin-A
| Macromolecule | Name: Glycophorin-A / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   human (human) human (human) |
| Molecular weight | Theoretical: 16.348433 KDa |
| Sequence | String: MYGKIIFVLL LSEIVSISAS STTGVAMHTS TSSSVTKSYI SSQTNDTHKR DTYAATPRAH EVSEISVRTV YPPEEETGER VQLAHHFSE PEITLIIFGV MAGVIGTILL ISYGIRRLIK KSPSDVKPLP SPDTDVPLSS VEIENPETSD Q |
-Macromolecule #5: Band 3 anion transport protein
| Macromolecule | Name: Band 3 anion transport protein / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   human (human) human (human) |
| Molecular weight | Theoretical: 101.883859 KDa |
| Sequence | String: MEELQDDYED MMEENLEQEE YEDPDIPESQ MEEPAAHDTE ATATDYHTTS HPGTHKVYVE LQELVMDEKN QELRWMEAAR WVQLEENLG ENGAWGRPHL SHLTFWSLLE LRRVFTKGTV LLDLQETSLA GVANQLLDRF IFEDQIRPQD REELLRALLL K HSHAGELE ...String: MEELQDDYED MMEENLEQEE YEDPDIPESQ MEEPAAHDTE ATATDYHTTS HPGTHKVYVE LQELVMDEKN QELRWMEAAR WVQLEENLG ENGAWGRPHL SHLTFWSLLE LRRVFTKGTV LLDLQETSLA GVANQLLDRF IFEDQIRPQD REELLRALLL K HSHAGELE ALGGVKPAVL TRSGDPSQPL LPQHSSLETQ LFCEQGDGGT EGHSPSGILE KIPPDSEATL VLVGRADFLE QP VLGFVRL QEAAELEAVE LPVPIRFLFV LLGPEAPHID YTQLGRAAAT LMSERVFRID AYMAQSRGEL LHSLEGFLDC SLV LPPTDA PSEQALLSLV PVQRELLRRR YQSSPAKPDS SFYKGLDLNG GPDDPLQQTG QLFGGLVRDI RRRYPYYLSD ITDA FSPQV LAAVIFIYFA ALSPAITFGG LLGEKTRNQM GVSELLISTA VQGILFALLG AQPLLVVGFS GPLLVFEEAF FSFCE TNGL EYIVGRVWIG FWLILLVVLV VAFEGSFLVR FISRYTQEIF SFLISLIFIY ETFSKLIKIF QDHPLQKTYN YNVLMV PKP QGPLPNTALL SLVLMAGTFF FAMMLRKFKN SSYFPGKLRR VIGDFGVPIS ILIMVLVDFF IQDTYTQKLS VPDGFKV SN SSARGWVIHP LGLRSEFPIW MMFASALPAL LVFILIFLES QITTLIVSKP ERKMVKGSGF HLDLLLVVGM GGVAALFG M PWLSATTVRS VTHANALTVM GKASTPGAAA QIQEVKEQRI SGLLVAVLVG LSILMEPILS RIPLAVLFGI FLYMGVTSL SGIQLFDRIL LLFKPPKYHP DVPYVKRVKT WRMHLFTGIQ IICLAVLWVV KSTPASLALP FVLILTVPLR RVLLPLIFRN VELQCLDAD DAKATFDEEE GRDEYDEVAM PV |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: Final gel filtration buffer contained 0.05% w/v digitonin, 130 mM KCl, 20 mM HEPES, pH 7.4, 1 mM ATP, 1 mM MgCl2, 1 mM PMSF. Peak fractions were concentrated to 8 mg/mL, and 0.01% w/v ...Details: Final gel filtration buffer contained 0.05% w/v digitonin, 130 mM KCl, 20 mM HEPES, pH 7.4, 1 mM ATP, 1 mM MgCl2, 1 mM PMSF. Peak fractions were concentrated to 8 mg/mL, and 0.01% w/v glycyrrhizic acid was added immediately prior to vitrification. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 4-6 seconds, wait time 30 seconds.. |
| Details | Ankyrin complex mixture purified from digitonin-solubilized erythrocyte ghost membranes |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 14464 / Average exposure time: 2.5 sec. / Average electron dose: 58.0 e/Å2 / Details: Two grids were imaged in a single session. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z
Z Y
Y X
X