[English] 日本語
 Yorodumi
Yorodumi- EMDB-16433: Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | |||||||||||||||||||||
 Map data Map data | Postprocess map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Sulfolactaldehyde dehydrogenase NADH GabD /  OXIDOREDUCTASE OXIDOREDUCTASE | |||||||||||||||||||||
| Biological species |  Rhizobium leguminosarum bv. trifolii SRDI565 (bacteria) Rhizobium leguminosarum bv. trifolii SRDI565 (bacteria) | |||||||||||||||||||||
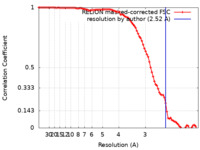
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.52 Å cryo EM / Resolution: 2.52 Å | |||||||||||||||||||||
 Authors Authors | Sharma M / Meek RW / Armstrong Z / Blaza JN / Alhifthi A / Li J / Goddard-Borger ED / Williams SJ / Davies GJ | |||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Australia, 6 items Australia, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Chem Sci / Year: 2023 Journal: Chem Sci / Year: 2023Title: Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from . Authors: Jinling Li / Mahima Sharma / Richard Meek / Amani Alhifthi / Zachary Armstrong / Niccolay Madiedo Soler / Mihwa Lee / Ethan D Goddard-Borger / James N Blaza / Gideon J Davies / Spencer J Williams /    Abstract: Sulfolactate (SL) is a short-chain organosulfonate that is an important reservoir of sulfur in the biosphere. SL is produced by oxidation of sulfolactaldehyde (SLA), which in turn derives from ...Sulfolactate (SL) is a short-chain organosulfonate that is an important reservoir of sulfur in the biosphere. SL is produced by oxidation of sulfolactaldehyde (SLA), which in turn derives from sulfoglycolysis of the sulfosugar sulfoquinovose, or through oxidation of 2,3-dihydroxypropanesulfonate. Oxidation of SLA is catalyzed by SLA dehydrogenases belonging to the aldehyde dehydrogenase superfamily. We report that SLA dehydrogenase GabD from the sulfoglycolytic bacterium SRDI565 can use both NAD and NADP as cofactor to oxidize SLA, and indicatively operates through a rapid equilibrium ordered mechanism. We report the cryo-EM structure of GabD bound to NADH, revealing a tetrameric quaternary structure and supporting proposal of organosulfonate binding residues in the active site, and a catalytic mechanism. Sequence based homology searches identified SLA dehydrogenase homologs in a range of putative sulfoglycolytic gene clusters in bacteria predominantly from the phyla Actinobacteria, Firmicutes, and Proteobacteria. This work provides a structural and biochemical view of SLA dehydrogenases to complement our knowledge of SLA reductases, and provide detailed insights into a critical step in the organosulfur cycle. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16433.map.gz emd_16433.map.gz | 60 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16433-v30.xml emd-16433-v30.xml emd-16433.xml emd-16433.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16433_fsc.xml emd_16433_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_16433.png emd_16433.png | 202.7 KB | ||
| Masks |  emd_16433_msk_1.map emd_16433_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16433.cif.gz emd-16433.cif.gz | 6.3 KB | ||
| Others |  emd_16433_half_map_1.map.gz emd_16433_half_map_1.map.gz emd_16433_half_map_2.map.gz emd_16433_half_map_2.map.gz | 48.4 MB 48.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16433 http://ftp.pdbj.org/pub/emdb/structures/EMD-16433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16433 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16433.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16433.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16433_msk_1.map emd_16433_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: HalfMap2
| File | emd_16433_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HalfMap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: HalfMap1
| File | emd_16433_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HalfMap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric assembly of RlGabD
| Entire | Name: Tetrameric assembly of RlGabD |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric assembly of RlGabD
| Supramolecule | Name: Tetrameric assembly of RlGabD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Rhizobium leguminosarum bv. trifolii SRDI565 (bacteria) Rhizobium leguminosarum bv. trifolii SRDI565 (bacteria) |
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: Succinate semialdehyde dehydrogenase
| Macromolecule | Name: Succinate semialdehyde dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rhizobium leguminosarum bv. trifolii SRDI565 (bacteria) Rhizobium leguminosarum bv. trifolii SRDI565 (bacteria) |
| Molecular weight | Theoretical: 52.697355 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MTERRTLALK DPALLTDKAF VAGAWIGAPD GKSIAVTDPF DGALIANVPD LGQAVVAQAI DAAAEAQKLW ERRTAKERAQ VLKRWYDLI VENADDLALI LTTEQGKPLA EARGEVVSNA AYLEWFAEEA KRIDGDIIPG PNAGQRIMVL KQPVGVCAAI T PWNFPNGM ...String: MTERRTLALK DPALLTDKAF VAGAWIGAPD GKSIAVTDPF DGALIANVPD LGQAVVAQAI DAAAEAQKLW ERRTAKERAQ VLKRWYDLI VENADDLALI LTTEQGKPLA EARGEVVSNA AYLEWFAEEA KRIDGDIIPG PNAGQRIMVL KQPVGVCAAI T PWNFPNGM ITRKAGPALA AGCSMILKPA SQTPLSALAL AVLAERAGVP KGVFSVVTGE AKPIGLEFCH NPRIAKITFT GS TGVGRWL MKEAAGGIKR LSLELGGNAP FIVFDDADLD AAVEGAMISK FRNAGQTCVC ANRIYVQESV AAAFTERLLA KVS GLSLGR GTDAGVSQGP LIDEKAVAKM EEHIADAMAN GGTVLTGGKR SVLGGTFFEP TVVTGVTQTM KVAKEETFAP LAPI ISFKH ENDVIAMAND SEFGLASYFY AKDMARIWRV AEALEAGMVG VNTGMIANEM APFGGIKQSG TGREGSKYGI EGFLE LKYV ALGGMLEHHH HHH |
-Macromolecule #2: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE
| Macromolecule | Name: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 4 / Formula: NAI |
|---|---|
| Molecular weight | Theoretical: 665.441 Da |
| Chemical component information |  ChemComp-NAI: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 330 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50mM Tris, 300mM NaCl, pH7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 240000 Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 240000 |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)