+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14196 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of human telomerase-DNA-TPP1 complex (sharpened) | |||||||||
 Map data Map data | Post-processed map of DNA-bound human telomerase in complex with TPP1 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation /  RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus / RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus /  telomerase activity ...positive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation / telomerase activity ...positive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation /  RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus / RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus /  telomerase activity / telomerase RNA reverse transcriptase activity / RNA-templated DNA biosynthetic process / establishment of protein localization to telomere / telomerase activity / telomerase RNA reverse transcriptase activity / RNA-templated DNA biosynthetic process / establishment of protein localization to telomere /  shelterin complex / nuclear telomere cap complex / siRNA processing / shelterin complex / nuclear telomere cap complex / siRNA processing /  telomerase RNA binding / telomere capping / telomerase RNA binding / telomere capping /  telomerase holoenzyme complex / positive regulation of vascular associated smooth muscle cell migration / telomeric DNA binding / DNA biosynthetic process / RNA-templated transcription / positive regulation of stem cell proliferation / mitochondrial nucleoid / negative regulation of cellular senescence / Telomere Extension By Telomerase / positive regulation of Wnt signaling pathway / telomere maintenance via telomerase / telomerase holoenzyme complex / positive regulation of vascular associated smooth muscle cell migration / telomeric DNA binding / DNA biosynthetic process / RNA-templated transcription / positive regulation of stem cell proliferation / mitochondrial nucleoid / negative regulation of cellular senescence / Telomere Extension By Telomerase / positive regulation of Wnt signaling pathway / telomere maintenance via telomerase /  replicative senescence / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of G1/S transition of mitotic cell cycle / response to cadmium ion / negative regulation of endothelial cell apoptotic process / positive regulation of telomerase activity / positive regulation of vascular associated smooth muscle cell proliferation / replicative senescence / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of G1/S transition of mitotic cell cycle / response to cadmium ion / negative regulation of endothelial cell apoptotic process / positive regulation of telomerase activity / positive regulation of vascular associated smooth muscle cell proliferation /  telomere maintenance / mitochondrion organization / positive regulation of nitric-oxide synthase activity / positive regulation of glucose import / Formation of the beta-catenin:TCF transactivating complex / telomere maintenance / mitochondrion organization / positive regulation of nitric-oxide synthase activity / positive regulation of glucose import / Formation of the beta-catenin:TCF transactivating complex /  regulation of protein stability / regulation of protein stability /  transcription coactivator binding / PML body / positive regulation of miRNA transcription / transcription coactivator binding / PML body / positive regulation of miRNA transcription /  RNA-directed DNA polymerase / structural constituent of chromatin / positive regulation of angiogenesis / RNA-directed DNA polymerase / structural constituent of chromatin / positive regulation of angiogenesis /  RNA-directed DNA polymerase activity / RNA-directed DNA polymerase activity /  nucleosome / positive regulation of protein binding / cellular response to hypoxia / protein-folding chaperone binding / negative regulation of neuron apoptotic process / nucleosome / positive regulation of protein binding / cellular response to hypoxia / protein-folding chaperone binding / negative regulation of neuron apoptotic process /  tRNA binding / tRNA binding /  chromosome, telomeric region / chromosome, telomeric region /  nuclear body / nuclear speck / protein heterodimerization activity / nuclear body / nuclear speck / protein heterodimerization activity /  RNA-dependent RNA polymerase activity / negative regulation of gene expression / RNA-dependent RNA polymerase activity / negative regulation of gene expression /  nucleolus / protein homodimerization activity / nucleolus / protein homodimerization activity /  DNA binding / DNA binding /  RNA binding / RNA binding /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   human (human) human (human) | |||||||||
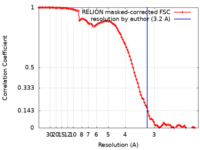
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Sekne Z / Ghanim GE / van Roon AMM / Nguyen THD | |||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structural basis of human telomerase recruitment by TPP1-POT1. Authors: Zala Sekne / George E Ghanim / Anne-Marie M van Roon / Thi Hoang Duong Nguyen /  Abstract: Telomerase maintains genome stability by extending the 3' telomeric repeats at eukaryotic chromosome ends, thereby counterbalancing progressive loss caused by incomplete genome replication. In ...Telomerase maintains genome stability by extending the 3' telomeric repeats at eukaryotic chromosome ends, thereby counterbalancing progressive loss caused by incomplete genome replication. In mammals, telomerase recruitment to telomeres is mediated by TPP1, which assembles as a heterodimer with POT1. We report structures of DNA-bound telomerase in complex with TPP1 and with TPP1-POT1 at 3.2- and 3.9-angstrom resolution, respectively. Our structures define interactions between telomerase and TPP1-POT1 that are crucial for telomerase recruitment to telomeres. The presence of TPP1-POT1 stabilizes the DNA, revealing an unexpected path by which DNA exits the telomerase active site and a DNA anchor site on telomerase that is important for telomerase processivity. Our findings rationalize extensive prior genetic and biochemical findings and provide a framework for future mechanistic work on telomerase regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14196.map.gz emd_14196.map.gz | 78.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14196-v30.xml emd-14196-v30.xml emd-14196.xml emd-14196.xml | 30 KB 30 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14196_fsc.xml emd_14196_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14196.png emd_14196.png | 110.7 KB | ||
| Masks |  emd_14196_msk_1.map emd_14196_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Others |  emd_14196_half_map_1.map.gz emd_14196_half_map_1.map.gz emd_14196_half_map_2.map.gz emd_14196_half_map_2.map.gz | 65.4 MB 65.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14196 http://ftp.pdbj.org/pub/emdb/structures/EMD-14196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14196 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14196 | HTTPS FTP |
-Related structure data
| Related structure data |  7qxaMC  7qxbC  7qxsC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14196.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14196.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map of DNA-bound human telomerase in complex with TPP1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
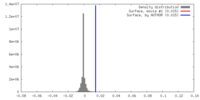
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_14196_msk_1.map emd_14196_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
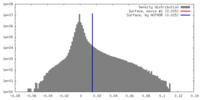
| Density Histograms |
-Half map: Half-map 2 from auto-refinement in RELION
| File | emd_14196_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 from auto-refinement in RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 from auto-refinement in RELION
| File | emd_14196_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 from auto-refinement in RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of telomeric DNA-bound human telomerase with TPP1
+Supramolecule #1: Complex of telomeric DNA-bound human telomerase with TPP1
+Supramolecule #2: Telomeric DNA
+Supramolecule #3: Telomerase reverse transcriptase and telomeric RNA
+Supramolecule #4: Histones
+Supramolecule #5: TPP1
+Macromolecule #1: Telomerase reverse transcriptase
+Macromolecule #3: Histone H2A
+Macromolecule #4: Histone H2B
+Macromolecule #6: Adrenocortical dysplasia homolog (Mouse), isoform CRA_a
+Macromolecule #2: human telomerase RNA
+Macromolecule #5: Telomeric DNA
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 45872 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 3 / Number real images: 50775 / Average exposure time: 3.0 sec. / Average electron dose: 48.0 e/Å2 Details: Images were collected in movie-mode and fractionated into 48 movie frames |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)