[English] 日本語
 Yorodumi
Yorodumi- EMDB-12174: The catalytic core lobe of human telomerase in complex with a tel... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12174 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
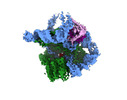
| Title | The catalytic core lobe of human telomerase in complex with a telomeric DNA substrate | |||||||||
 Map data Map data | Post-processed map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation /  RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus / RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus /  telomerase activity ...positive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation / telomerase activity ...positive regulation of hair cycle / template-free RNA nucleotidyltransferase / positive regulation of transdifferentiation / TERT-RMRP complex / DNA strand elongation /  RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus / RNA-directed RNA polymerase complex / telomerase catalytic core complex / siRNA transcription / positive regulation of protein localization to nucleolus /  telomerase activity / telomerase RNA reverse transcriptase activity / RNA-templated DNA biosynthetic process / establishment of protein localization to telomere / nuclear telomere cap complex / siRNA processing / telomerase activity / telomerase RNA reverse transcriptase activity / RNA-templated DNA biosynthetic process / establishment of protein localization to telomere / nuclear telomere cap complex / siRNA processing /  telomerase RNA binding / telomerase RNA binding /  telomerase holoenzyme complex / positive regulation of vascular associated smooth muscle cell migration / telomeric DNA binding / DNA biosynthetic process / RNA-templated transcription / positive regulation of stem cell proliferation / mitochondrial nucleoid / negative regulation of cellular senescence / Telomere Extension By Telomerase / positive regulation of Wnt signaling pathway / telomere maintenance via telomerase / telomerase holoenzyme complex / positive regulation of vascular associated smooth muscle cell migration / telomeric DNA binding / DNA biosynthetic process / RNA-templated transcription / positive regulation of stem cell proliferation / mitochondrial nucleoid / negative regulation of cellular senescence / Telomere Extension By Telomerase / positive regulation of Wnt signaling pathway / telomere maintenance via telomerase /  replicative senescence / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of G1/S transition of mitotic cell cycle / response to cadmium ion / negative regulation of endothelial cell apoptotic process / positive regulation of vascular associated smooth muscle cell proliferation / replicative senescence / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of G1/S transition of mitotic cell cycle / response to cadmium ion / negative regulation of endothelial cell apoptotic process / positive regulation of vascular associated smooth muscle cell proliferation /  telomere maintenance / mitochondrion organization / positive regulation of nitric-oxide synthase activity / positive regulation of glucose import / Formation of the beta-catenin:TCF transactivating complex / telomere maintenance / mitochondrion organization / positive regulation of nitric-oxide synthase activity / positive regulation of glucose import / Formation of the beta-catenin:TCF transactivating complex /  regulation of protein stability / regulation of protein stability /  transcription coactivator binding / PML body / positive regulation of miRNA transcription / structural constituent of chromatin / transcription coactivator binding / PML body / positive regulation of miRNA transcription / structural constituent of chromatin /  RNA-directed DNA polymerase / positive regulation of angiogenesis / RNA-directed DNA polymerase / positive regulation of angiogenesis /  RNA-directed DNA polymerase activity / RNA-directed DNA polymerase activity /  nucleosome / positive regulation of protein binding / cellular response to hypoxia / protein-folding chaperone binding / negative regulation of neuron apoptotic process / nucleosome / positive regulation of protein binding / cellular response to hypoxia / protein-folding chaperone binding / negative regulation of neuron apoptotic process /  tRNA binding / tRNA binding /  chromosome, telomeric region / nuclear speck / protein heterodimerization activity / chromosome, telomeric region / nuclear speck / protein heterodimerization activity /  RNA-dependent RNA polymerase activity / negative regulation of gene expression / RNA-dependent RNA polymerase activity / negative regulation of gene expression /  nucleolus / protein homodimerization activity / nucleolus / protein homodimerization activity /  DNA binding / DNA binding /  RNA binding / RNA binding /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) / Homo sapiens (human) / synthetic construct (others) /   Human (human) Human (human) | |||||||||
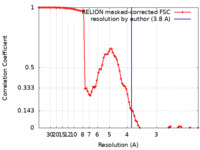
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Nguyen THD / Ghanim GE / Fountain AJ / van Roon AMM / Rangan R / Das R / Collins K | |||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure of human telomerase holoenzyme with bound telomeric DNA. Authors: George E Ghanim / Adam J Fountain / Anne-Marie M van Roon / Ramya Rangan / Rhiju Das / Kathleen Collins / Thi Hoang Duong Nguyen /   Abstract: Telomerase adds telomeric repeats at chromosome ends to compensate for the telomere loss that is caused by incomplete genome end replication. In humans, telomerase is upregulated during embryogenesis ...Telomerase adds telomeric repeats at chromosome ends to compensate for the telomere loss that is caused by incomplete genome end replication. In humans, telomerase is upregulated during embryogenesis and in cancers, and mutations that compromise the function of telomerase result in disease. A previous structure of human telomerase at a resolution of 8 Å revealed a vertebrate-specific composition and architecture, comprising a catalytic core that is flexibly tethered to an H and ACA (hereafter, H/ACA) box ribonucleoprotein (RNP) lobe by telomerase RNA. High-resolution structural information is necessary to develop treatments that can effectively modulate telomerase activity as a therapeutic approach against cancers and disease. Here we used cryo-electron microscopy to determine the structure of human telomerase holoenzyme bound to telomeric DNA at sub-4 Å resolution, which reveals crucial DNA- and RNA-binding interfaces in the active site of telomerase as well as the locations of mutations that alter telomerase activity. We identified a histone H2A-H2B dimer within the holoenzyme that was bound to an essential telomerase RNA motif, which suggests a role for histones in the folding and function of telomerase RNA. Furthermore, this structure of a eukaryotic H/ACA RNP reveals the molecular recognition of conserved RNA and protein motifs, as well as interactions that are crucial for understanding the molecular pathology of many mutations that cause disease. Our findings provide the structural details of the assembly and active site of human telomerase, which paves the way for the development of therapeutic agents that target this enzyme. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12174.map.gz emd_12174.map.gz | 48.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12174-v30.xml emd-12174-v30.xml emd-12174.xml emd-12174.xml | 27.4 KB 27.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12174_fsc.xml emd_12174_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12174.png emd_12174.png | 109.9 KB | ||
| Others |  emd_12174_half_map_1.map.gz emd_12174_half_map_1.map.gz emd_12174_half_map_2.map.gz emd_12174_half_map_2.map.gz | 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12174 http://ftp.pdbj.org/pub/emdb/structures/EMD-12174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12174 | HTTPS FTP |
-Related structure data
| Related structure data |  7bg9MC  7bgbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10732 (Title: Structure of human telomerase holoenzyme with bound telomeric DNA EMPIAR-10732 (Title: Structure of human telomerase holoenzyme with bound telomeric DNAData size: 11.2 TB Data #1: Unaligned multiframe micrographs of human telomerase holoenzyme bound to a telomeric DNA [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12174.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12174.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
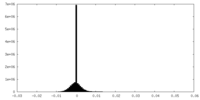
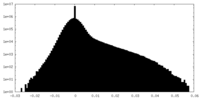
-Half map: #2
| File | emd_12174_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
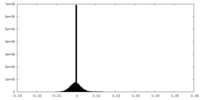
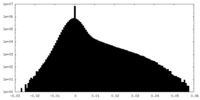
-Half map: #1
| File | emd_12174_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human telomerase catalytic core in complex with a telomeric DNA s...
| Entire | Name: Human telomerase catalytic core in complex with a telomeric DNA substrate |
|---|---|
| Components |
|
-Supramolecule #1: Human telomerase catalytic core in complex with a telomeric DNA s...
| Supramolecule | Name: Human telomerase catalytic core in complex with a telomeric DNA substrate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: Telomerase reverse transcriptase
| Supramolecule | Name: Telomerase reverse transcriptase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Histone
| Supramolecule | Name: Histone / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: DNA
| Supramolecule | Name: DNA / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
-Macromolecule #1: Telomerase reverse transcriptase,Telomerase reverse transcriptase
| Macromolecule | Name: Telomerase reverse transcriptase,Telomerase reverse transcriptase type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  RNA-directed DNA polymerase RNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Organ: Kidney Homo sapiens (human) / Organ: Kidney |
| Molecular weight | Theoretical: 149.158578 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTAALAQHD EAVDNKFNKE QQNAFYEILH LPNLNEEQRN AFIQSLKDDP SQSANLLAEA KKLNDAQAPK VDNKFNKEQQ NAFYEILHL PNLNEEQRNA FIQSLKDDPS QSANLLAEAK KLNGAQAPKV DANSAGKSTD YDIPTTASEN LYFQGHKLGT F QGPWSHPQ ...String: MKTAALAQHD EAVDNKFNKE QQNAFYEILH LPNLNEEQRN AFIQSLKDDP SQSANLLAEA KKLNDAQAPK VDNKFNKEQQ NAFYEILHL PNLNEEQRNA FIQSLKDDPS QSANLLAEAK KLNGAQAPKV DANSAGKSTD YDIPTTASEN LYFQGHKLGT F QGPWSHPQ FEKGSAGSAA GSGAGWSHPQ FEKSRPTTAS GTMPRAPRCR AVRSLLRSHY REVLPLATFV RRLGPQGWRL VQ RGDPAAF RALVAQCLVC VPWDARPPPA APSFRQVSCL KELVARVLQR LCERGAKNVL AFGFALLDGA RGGPPEAFTT SVR SYLPNT VTDALRGSGA WGLLLRRVGD DVLVHLLARC ALFVLVAPSC AYQVCGPPLY QLGAATQARP PPHASGPRRR LGCE RAWNH SVREAGVPLG LPAPGARRRG GSASRSLPLP KRPRRGAAPE PERTPVGQGS WAHPGRTRGP SDRGFCVVSP ARPAE EATS LEGALSGTRH SHPSVGRQHH AGPPSTSRPP RPWDTPCPPV YAETKHFLYS SGDKEQLRPS FLLSSLRPSL TGARRL VET IFLGSRPWMP GTPRRLPRLP QRYWQMRPLF LELLGNHAQC PYGVLLKTHC PLRAAVTPAA GVCAREKPQG SVAAPEE ED TDPRRLVQLL RQHSSPWQVY GFVRACLRRL VPPGLWGSRH NERRFLRNTK KFISLGKHAK LSLQELTWKM SVRDCAWL R RSPGVGCVPA AEHRLREEIL AKFLHWLMSV YVVELLRSFF YVTETTFQKN RLFFYRKSVW SKLQSIGIRQ HLKRVQLRE LSEAEVRQHR EARPALLTSR LRFIPKPDGL RPIVNMDYVV GARTFRREKR AERLTSRVKA LFSVLNYERA RRPGLLGASV LGLDDIHRA WRTFVLRVRA QDPPPELYFV KVDVTGAYDT IPQDRLTEVI ASIIKPQNTY CVRRYAVVQK AAHGHVRKAF K SHVSTLTD LQPYMRQFVA HLQETSPLRD AVVIEQSSSL NEASSGLFDV FLRFMCHHAV RIRGKSYVQC QGIPQGSILS TL LCSLCYG DMENKLFAGI RRDGLLLRLV DDFLLVTPHL THAKTFLRTL VRGVPEYGCV VNLRKTVVNF PVEDEALGGT AFV QMPAHG LFPWCGLLLD TRTLEVQSDY SSYARTSIRA SLTFNRGFKA GRNMRRKLFG VLRLKCHSLF LDLQVNSLQT VCTN IYKIL LLQAYRFHAC VLQLPFHQQV WKNPTFFLRV ISDTASLCYS ILKAKNAGMS LGAKGAAGPL PSEAVQWLCH QAFLL KLTR HRVTYVPLLG SLRTAQTQLS RKLPGTTLTA LEAAANPALP SDFKTILD |
-Macromolecule #3: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human (human) / Organ: Kidney Human (human) / Organ: Kidney |
| Molecular weight | Theoretical: 18.074932 KDa |
| Sequence | String: MPDPAKSAPA PKKGSKKAVT KVQKKDGKKR KRSRKESYSV YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV TKYTSSNPRN LSPTKPGGSE DRQPPPSQLS AIPPFCLVLR A GIAGQV |
-Macromolecule #4: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human (human) / Organ: Kidney Human (human) / Organ: Kidney |
| Molecular weight | Theoretical: 14.140584 KDa |
| Sequence | String: MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VRRLLRKGNY AERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG KVTIAQGGVL PNIQAVLLPK KTESHHKAKG K |
-Macromolecule #2: DNA (5'-D(P*TP*TP*AP*GP*GP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*AP*GP*GP*G)-3') / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 5.514567 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DA)(DG)(DG)(DG) |
-Macromolecule #5: RNA (256-MER)
| Macromolecule | Name: RNA (256-MER) / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Organ: Kidney Homo sapiens (human) / Organ: Kidney |
| Molecular weight | Theoretical: 145.477797 KDa |
| Sequence | String: GGGUUGCGGA GGGUGGGCCU GGGAGGGGUG GUGGCCAUUU UUUGUCUAAC CCUAACUGAG AAGGGCGUAG GCGCCGUGCU UUUGCUCCC CGCGCGCUGU UUUUCUCGCU GACUUUCAGC GGGCGGAAAA GCCUCGGCCU GCCGCCUUCC ACCGUUCAUU C UAGAGCAA ...String: GGGUUGCGGA GGGUGGGCCU GGGAGGGGUG GUGGCCAUUU UUUGUCUAAC CCUAACUGAG AAGGGCGUAG GCGCCGUGCU UUUGCUCCC CGCGCGCUGU UUUUCUCGCU GACUUUCAGC GGGCGGAAAA GCCUCGGCCU GCCGCCUUCC ACCGUUCAUU C UAGAGCAA ACAAAAAAUG UCAGCUGCUG GCCCGUUCGC CCCUCCCGGG GACCUGCGGC GGGUCGCCUG CCCAGCCCCC GA ACCCCGC CUGGAGGCCG CGGUCGGCCC GGGGCUUCUC CGGAGGCACC CACUGCCACC GCGAAGAGUU GGGCUCUGUC AGC CGCGGG UCUCUCGGGG GCGAGGGCGA GGUUCAGGCC UUUCAGGCCG CAGGAAGAGG AACGGAGCGA GUCCCCGCGC GCGG CGCGA UUCCCUGAGC UGUGGGACGU GCACCCAGGA CUCGGCUCAC ACAUGC |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4-5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 78.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 43639 / Average exposure time: 1.0 sec. / Average electron dose: 47.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 80 |
|---|---|
| Output model |  PDB-7bg9: |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X