[English] 日本語
 Yorodumi
Yorodumi- EMDB-13144: Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13144 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationintracellular monoatomic cation homeostasis / xenobiotic detoxification by transmembrane export across the plasma membrane / ABC-type xenobiotic transporter activity / response to cycloheximide / cell periphery / response to xenobiotic stimulus / response to antibiotic /  ATP hydrolysis activity / ATP hydrolysis activity /  mitochondrion / mitochondrion /  ATP binding ...intracellular monoatomic cation homeostasis / xenobiotic detoxification by transmembrane export across the plasma membrane / ABC-type xenobiotic transporter activity / response to cycloheximide / cell periphery / response to xenobiotic stimulus / response to antibiotic / ATP binding ...intracellular monoatomic cation homeostasis / xenobiotic detoxification by transmembrane export across the plasma membrane / ABC-type xenobiotic transporter activity / response to cycloheximide / cell periphery / response to xenobiotic stimulus / response to antibiotic /  ATP hydrolysis activity / ATP hydrolysis activity /  mitochondrion / mitochondrion /  ATP binding / identical protein binding / ATP binding / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.13 Å cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Szewczak-Harris A / Wagner M / Du D / Schmitt L / Luisi BF | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Authors: Andrzej Harris / Manuel Wagner / Dijun Du / Stefanie Raschka / Lea-Marie Nentwig / Holger Gohlke / Sander H J Smits / Ben F Luisi / Lutz Schmitt /    Abstract: Pdr5, a member of the extensive ABC transporter superfamily, is representative of a clinically relevant subgroup involved in pleiotropic drug resistance. Pdr5 and its homologues drive drug efflux ...Pdr5, a member of the extensive ABC transporter superfamily, is representative of a clinically relevant subgroup involved in pleiotropic drug resistance. Pdr5 and its homologues drive drug efflux through uncoupled hydrolysis of nucleotides, enabling organisms such as baker's yeast and pathogenic fungi to survive in the presence of chemically diverse antifungal agents. Here, we present the molecular structure of Pdr5 solved with single particle cryo-EM, revealing details of an ATP-driven conformational cycle, which mechanically drives drug translocation through an amphipathic channel, and a clamping switch within a conserved linker loop that acts as a nucleotide sensor. One half of the transporter remains nearly invariant throughout the cycle, while its partner undergoes changes that are transmitted across inter-domain interfaces to support a peristaltic motion of the pumped molecule. The efflux model proposed here rationalises the pleiotropic impact of Pdr5 and opens new avenues for the development of effective antifungal compounds. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13144.map.gz emd_13144.map.gz | 500.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13144-v30.xml emd-13144-v30.xml emd-13144.xml emd-13144.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13144.png emd_13144.png | 49.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13144 http://ftp.pdbj.org/pub/emdb/structures/EMD-13144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13144 | HTTPS FTP |
-Related structure data
| Related structure data |  7p05MC  7p03C  7p04C  7p06C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13144.map.gz / Format: CCP4 / Size: 548.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13144.map.gz / Format: CCP4 / Size: 548.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.652 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward...
| Entire | Name: Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward...
| Supramolecule | Name: Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast)Location in cell: cell membrane |
-Macromolecule #1: Pleiotropic ABC efflux transporter of multiple drugs
| Macromolecule | Name: Pleiotropic ABC efflux transporter of multiple drugs / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Molecular weight | Theoretical: 170.61725 KDa |
| Sequence | String: MPEAKLNNNV NDVTSYSSAS SSTENAADLH NYNGFDEHTE ARIQKLARTL TAQSMQNSTQ SAPNKSDAQS IFSSGVEGVN PIFSDPEAP GYDPKLDPNS ENFSSAAWVK NMAHLSAADP DFYKPYSLGC AWKNLSASGA SADVAYQSTV VNIPYKILKS G LRKFQRSK ...String: MPEAKLNNNV NDVTSYSSAS SSTENAADLH NYNGFDEHTE ARIQKLARTL TAQSMQNSTQ SAPNKSDAQS IFSSGVEGVN PIFSDPEAP GYDPKLDPNS ENFSSAAWVK NMAHLSAADP DFYKPYSLGC AWKNLSASGA SADVAYQSTV VNIPYKILKS G LRKFQRSK ETNTFQILKP MDGCLNPGEL LVVLGRPGSG CTTLLKSISS NTHGFDLGAD TKISYSGYSG DDIKKHFRGE VV YNAEADV HLPHLTVFET LVTVARLKTP QNRIKGVDRE SYANHLAEVA MATYGLSHTR NTKVGNDIVR GVSGGERKRV SIA EVSICG SKFQCWDNAT RGLDSATALE FIRALKTQAD ISNTSATVAI YQCSQDAYDL FNKVCVLDDG YQIYYGPADK AKKY FEDMG YVCPSRQTTA DFLTSVTSPS ERTLNKDMLK KGIHIPQTPK EMNDYWVKSP NYKELMKEVD QRLLNDDEAS REAIK EAHI AKQSKRARPS SPYTVSYMMQ VKYLLIRNMW RLRNNIGFTL FMILGNCSMA LILGSMFFKI MKKGDTSTFY FRGSAM FFA ILFNAFSSLL EIFSLYEARP ITEKHRTYSL YHPSADAFAS VLSEIPSKLI IAVCFNIIFY FLVDFRRNGG VFFFYLL IN IVAVFSMSHL FRCVGSLTKT LSEAMVPASM LLLALSMYTG FAIPKKKILR WSKWIWYINP LAYLFESLLI NEFHGIKF P CAEYVPRGPA YANISSTESV CTVVGAVPGQ DYVLGDDFIR GTYQYYHKDK WRGFGIGMAY VVFFFFVYLF LCEYNEGAK QKGEILVFPR SIVKRMKKRG VLTEKNANDP ENVGERSDLS SDRKMLQESS EEESDTYGEI GLSKSEAIFH WRNLCYEVQI KAETRRILN NVDGWVKPGT LTALMGASGA GKTTLLDCLA ERVTMGVITG DILVNGIPRD KSFPRSIGYC QQQDLHLKTA T VRESLRFS AYLRQPAEVS IEEKNRYVEE VIKILEMEKY ADAVVGVAGE GLNVEQRKRL TIGVELTAKP KLLVFLDEPT SG LDSQTAW SICQLMKKLA NHGQAILCTI HQPSAILMQE FDRLLFMQRG GKTVYFGDLG EGCKTMIDYF ESHGAHKCPA DAN PAEWML EVVGAAPGSH ANQDYYEVWR NSEEYRAVQS ELDWMERELP KKGSITAAED KHEFSQSIIY QTKLVSIRLF QQYW RSPDY LWSKFILTIF NQLFIGFTFF KAGTSLQGLQ NQMLAVFMFT VIFNPILQQY LPSFVQQRDL YEARERPSRT FSWIS FIFA QIFVEVPWNI LAGTIAYFIY YYPIGFYSNA SAAGQLHERG ALFWLFSCAF YVYVGSMGLL VISFNQVAES AANLAS LLF TMSLSFCGVM TTPSAMPRFW IFMYRVSPLT YFIQALLAVG VANVDVKCAD YELLEFTPPS GMTCGQYMEP YLQLAKT GY LTDENATDTC SFCQISTTND YLANVNSFYS ERWRNYGIFI CYIAFNYIAG VFFYWLARVP KKNGKLSKK |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: RHODAMINE 6G
| Macromolecule | Name: RHODAMINE 6G / type: ligand / ID: 4 / Number of copies: 1 / Formula: RHQ |
|---|---|
| Molecular weight | Theoretical: 443.557 Da |
| Chemical component information | 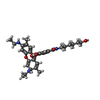 ChemComp-R6G: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 |
| Specialist optics | Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: HELIUM |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 1.31 sec. / Average electron dose: 47.73 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 4.1) |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final 3D classification | Software - Name: RELION (ver. 3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final reconstruction | Number classes used: 1 / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.13 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 76419 |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7p05: |
 Movie
Movie Controller
Controller


















