[English] 日本語
 Yorodumi
Yorodumi- PDB-7w72: Structure of a human glycosylphosphatidylinositol (GPI) transamidase -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7w72 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a human glycosylphosphatidylinositol (GPI) transamidase | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  glycosylphosphatidylinositol / transamidase glycosylphosphatidylinositol / transamidase | |||||||||
| Function / homology |  Function and homology information Function and homology informationGPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchor biosynthetic process / protein retention in ER lumen / Attachment of GPI anchor to uPAR /  Hydrolases / regulation of receptor signaling pathway via JAK-STAT / Hydrolases / regulation of receptor signaling pathway via JAK-STAT /  protein disulfide isomerase activity / protein disulfide isomerase activity /  tubulin binding ...GPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchor biosynthetic process / protein retention in ER lumen / Attachment of GPI anchor to uPAR / tubulin binding ...GPI-anchor transamidase activity / attachment of GPI anchor to protein / GPI-anchor transamidase complex / GPI anchor biosynthetic process / protein retention in ER lumen / Attachment of GPI anchor to uPAR /  Hydrolases / regulation of receptor signaling pathway via JAK-STAT / Hydrolases / regulation of receptor signaling pathway via JAK-STAT /  protein disulfide isomerase activity / protein disulfide isomerase activity /  tubulin binding / neuron differentiation / cytoplasmic vesicle / protein-containing complex assembly / neuron apoptotic process / tubulin binding / neuron differentiation / cytoplasmic vesicle / protein-containing complex assembly / neuron apoptotic process /  centrosome / endoplasmic reticulum membrane / centrosome / endoplasmic reticulum membrane /  endoplasmic reticulum / endoplasmic reticulum /  mitochondrion / mitochondrion /  proteolysis / proteolysis /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zhang, H. / Su, J. / Li, B. / Gao, Y. / Zhang, X.C. / Zhao, Y. | |||||||||
| Funding support |  China, 2items China, 2items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure of human glycosylphosphatidylinositol transamidase. Authors: Hongwei Zhang / Jiawei Su / Bin Li / Yiwei Gao / Mengran Liu / Lingli He / Hao Xu / Yanli Dong / Xuejun Cai Zhang / Yan Zhao /  Abstract: Glycosylphosphatidylinositol (GPI) molecules are complex glycophospholipids and serve as membrane anchors for tethering many proteins to the cell surface. Attaching GPI to the protein in the ...Glycosylphosphatidylinositol (GPI) molecules are complex glycophospholipids and serve as membrane anchors for tethering many proteins to the cell surface. Attaching GPI to the protein in the endoplasmic reticulum (ER) is catalyzed by the transmembrane GPI transamidase (GPIT) complex, which is essential for maturation of the GPI-anchored proteins. The GPIT complex is known to be composed of five subunits: PIGK, PIGU, PIGT, PIGS and GPAA1. Here, we determined the structure of the human GPIT complex at a resolution of 3.1 Å using single-particle cryo-EM, elucidating its overall assembly. The PIGK subunit functions as the catalytic component, in which we identified a C206-H164-N58 triad that is critical for the transamination reaction. Transmembrane helices constitute a widely opened cleft, which is located underneath PIGK, serving as a GPI substrate-binding site. The ubiquitin E3 ligase RNF121 is visualized at the back of the complex and probably serves as a quality control factor for the GPIT complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7w72.cif.gz 7w72.cif.gz | 429.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7w72.ent.gz pdb7w72.ent.gz | 352.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7w72.json.gz 7w72.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/w7/7w72 https://data.pdbj.org/pub/pdb/validation_reports/w7/7w72 ftp://data.pdbj.org/pub/pdb/validation_reports/w7/7w72 ftp://data.pdbj.org/pub/pdb/validation_reports/w7/7w72 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  32336MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 3 types, 3 molecules UKA
| #1: Protein | Mass: 48448.422 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PIGU / Cell line (production host): HEK293 / Production host: Homo sapiens (human) / Gene: PIGU / Cell line (production host): HEK293 / Production host:   Homo sapiens (human) / References: UniProt: Q9H490 Homo sapiens (human) / References: UniProt: Q9H490 |
|---|---|
| #4: Protein | Mass: 45302.629 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PIGK / Cell line (production host): HEK293 / Production host: Homo sapiens (human) / Gene: PIGK / Cell line (production host): HEK293 / Production host:   Homo sapiens (human) / References: UniProt: Q92643, Homo sapiens (human) / References: UniProt: Q92643,  Hydrolases Hydrolases |
| #5: Protein | Mass: 67683.836 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: GPAA1, GAA1 / Cell line (production host): HEK293 / Production host: Homo sapiens (human) / Gene: GPAA1, GAA1 / Cell line (production host): HEK293 / Production host:   Homo sapiens (human) / References: UniProt: O43292 Homo sapiens (human) / References: UniProt: O43292 |
-GPI transamidase component PIG- ... , 2 types, 2 molecules ST
| #2: Protein | Mass: 59097.820 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PIGS / Cell line (production host): HEK293 / Production host: Homo sapiens (human) / Gene: PIGS / Cell line (production host): HEK293 / Production host:   Homo sapiens (human) / References: UniProt: Q96S52 Homo sapiens (human) / References: UniProt: Q96S52 |
|---|---|
| #3: Protein | Mass: 60397.691 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PIGT / Cell line (production host): HEK293 / Production host: Homo sapiens (human) / Gene: PIGT / Cell line (production host): HEK293 / Production host:   Homo sapiens (human) / References: UniProt: Q969N2 Homo sapiens (human) / References: UniProt: Q969N2 |
-Sugars , 2 types, 3 molecules 
| #6: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose / Mass: 586.542 Da / Num. of mol.: 1 / Source method: obtained synthetically / Mass: 586.542 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #8: Sugar |  N-Acetylglucosamine N-Acetylglucosamine |
-Non-polymers , 2 types, 2 molecules 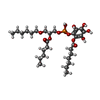


| #7: Chemical | ChemComp-8JY / [ |
|---|---|
| #9: Chemical | ChemComp-CA / |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Human glycosylphosphatidylinositol transamidase / Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:   Homo sapiens (human) / Cell: HEK293 Homo sapiens (human) / Cell: HEK293 |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 22000 nm / Nominal defocus min: 12000 nm Bright-field microscopy / Nominal defocus max: 22000 nm / Nominal defocus min: 12000 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 61148 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj



