| Entry | Database: PDB / ID: 5muc
|
|---|
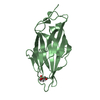
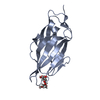
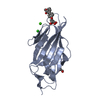
| Title | Crystal structure of the FimH lectin domain in complex with 1,5-Anhydromannitol |
|---|
 Components Components | Protein FimH |
|---|
 Keywords Keywords | SUGAR BINDING PROTEIN / FimH / UTI /  lectin / lectin /  carbohydrate / UPEC / carbohydrate / UPEC /  bladder infection bladder infection |
|---|
| Function / homology |  Function and homology information Function and homology information |
|---|
| Biological species |    Escherichia coli (E. coli) Escherichia coli (E. coli) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å |
|---|
 Authors Authors | Jakob, R.P. / Rabbani, S. / Ernst, B. / Maier, T. |
|---|
 Citation Citation |  Journal: Chemistry / Year: 2018 Journal: Chemistry / Year: 2018
Title: KinITC-One Method Supports both Thermodynamic and Kinetic SARs as Exemplified on FimH Antagonists.
Authors: Zihlmann, P. / Silbermann, M. / Sharpe, T. / Jiang, X. / Muhlethaler, T. / Jakob, R.P. / Rabbani, S. / Sager, C.P. / Frei, P. / Pang, L. / Maier, T. / Ernst, B. |
|---|
| History | | Deposition | Jan 13, 2017 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Feb 14, 2018 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 4, 2018 | Group: Data collection / Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.name |
|---|
| Revision 1.2 | Aug 8, 2018 | Group: Data collection / Database references / Category: citation / citation_author / Item: _citation.title |
|---|
| Revision 1.3 | Sep 12, 2018 | Group: Data collection / Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first / _citation.page_last |
|---|
| Revision 2.0 | Jul 29, 2020 | Group: Atomic model / Derived calculations / Structure summary
Category: atom_site / chem_comp ...atom_site / chem_comp / entity / pdbx_entity_nonpoly / struct_site / struct_site_gen
Item: _atom_site.auth_atom_id / _atom_site.label_atom_id ..._atom_site.auth_atom_id / _atom_site.label_atom_id / _chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 2.1 | Jan 17, 2024 | Group: Data collection / Database references ...Data collection / Database references / Refinement description / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords lectin /
lectin /  carbohydrate / UPEC /
carbohydrate / UPEC /  bladder infection
bladder infection Function and homology information
Function and homology information pilus / cell-substrate adhesion /
pilus / cell-substrate adhesion /  D-mannose binding / host cell membrane /
D-mannose binding / host cell membrane /  cell adhesion
cell adhesion
 Escherichia coli (E. coli)
Escherichia coli (E. coli) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å
MOLECULAR REPLACEMENT / Resolution: 2.6 Å  Authors
Authors Citation
Citation Journal: Chemistry / Year: 2018
Journal: Chemistry / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5muc.cif.gz
5muc.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5muc.ent.gz
pdb5muc.ent.gz PDB format
PDB format 5muc.json.gz
5muc.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/mu/5muc
https://data.pdbj.org/pub/pdb/validation_reports/mu/5muc ftp://data.pdbj.org/pub/pdb/validation_reports/mu/5muc
ftp://data.pdbj.org/pub/pdb/validation_reports/mu/5muc
 Links
Links Assembly
Assembly

 Components
Components
 Escherichia coli (strain K12) (bacteria)
Escherichia coli (strain K12) (bacteria)
 Escherichia coli BL21(DE3) (bacteria) / References: UniProt: P08191
Escherichia coli BL21(DE3) (bacteria) / References: UniProt: P08191 Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06SA / Wavelength: 1.87287 Å
/ Beamline: X06SA / Wavelength: 1.87287 Å : 1.87287 Å / Relative weight: 1
: 1.87287 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller



















 PDBj
PDBj