+Search query
-Structure paper
| Title | Structural basis for the ligand recognition and signaling of free fatty acid receptors. |
|---|---|
| Journal, issue, pages | Sci Adv, Vol. 10, Issue 2, Page eadj2384, Year 2024 |
| Publish date | Jan 12, 2024 |
 Authors Authors | Xuan Zhang / Abdul-Akim Guseinov / Laura Jenkins / Kunpeng Li / Irina G Tikhonova / Graeme Milligan / Cheng Zhang /   |
| PubMed Abstract | Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 ...Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 are activated by long-chain fatty acids, while FFA2 and FFA3 respond to short-chain fatty acids generated by intestinal microbiota. FFA1, FFA2, and FFA4 are potential drug targets for metabolic and inflammatory conditions. Here, we determined the active structures of FFA1 and FFA4 bound to docosahexaenoic acid, FFA4 bound to the synthetic agonist TUG-891, and butyrate-bound FFA2, each complexed with an engineered heterotrimeric G protein (miniG), by cryo-electron microscopy. Together with computational simulations and mutagenesis studies, we elucidated the similarities and differences in the binding modes of fatty acid ligands to their respective GPCRs. Our findings unveiled distinct mechanisms of receptor activation and G protein coupling. We anticipate that these outcomes will facilitate structure-based drug development and underpin future research on this group of GPCRs. |
 External links External links |  Sci Adv / Sci Adv /  PubMed:38198545 / PubMed:38198545 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.06 - 3.6 Å |
| Structure data | EMDB-41007, PDB-8t3o: EMDB-41008, PDB-8t3q: EMDB-41010, PDB-8t3s: EMDB-41013, PDB-8t3v:  EMDB-41014: Cryo-EM structure of the DHA bound FFA1-Gq complex(mask on receptor) |
| Chemicals | 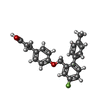 ChemComp-YN9:  ChemComp-2Y5:  ChemComp-PLM: 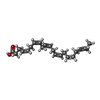 ChemComp-HXA:  ChemComp-BUA:  ChemComp-CLR: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  GPCR / GPCR /  Complex / Complex /  agonist agonist |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













