[English] 日本語
 Yorodumi
Yorodumi- EMDB-41014: Cryo-EM structure of the DHA bound FFA1-Gq complex(mask on receptor) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the DHA bound FFA1-Gq complex(mask on receptor) | |||||||||
 Map data Map data | DHA-FFA1-miniGq-Local | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  GPCR / GPCR /  Complex / Complex /  agonist / agonist /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Zhang X / Tikhonova I / Milligan G / Zhang C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Structural basis for the ligand recognition and signaling of free fatty acid receptors. Authors: Xuan Zhang / Abdul-Akim Guseinov / Laura Jenkins / Kunpeng Li / Irina G Tikhonova / Graeme Milligan / Cheng Zhang /   Abstract: Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 ...Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 are activated by long-chain fatty acids, while FFA2 and FFA3 respond to short-chain fatty acids generated by intestinal microbiota. FFA1, FFA2, and FFA4 are potential drug targets for metabolic and inflammatory conditions. Here, we determined the active structures of FFA1 and FFA4 bound to docosahexaenoic acid, FFA4 bound to the synthetic agonist TUG-891, and butyrate-bound FFA2, each complexed with an engineered heterotrimeric G protein (miniG), by cryo-electron microscopy. Together with computational simulations and mutagenesis studies, we elucidated the similarities and differences in the binding modes of fatty acid ligands to their respective GPCRs. Our findings unveiled distinct mechanisms of receptor activation and G protein coupling. We anticipate that these outcomes will facilitate structure-based drug development and underpin future research on this group of GPCRs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41014.map.gz emd_41014.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41014-v30.xml emd-41014-v30.xml emd-41014.xml emd-41014.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41014.png emd_41014.png | 17.1 KB | ||
| Filedesc metadata |  emd-41014.cif.gz emd-41014.cif.gz | 3.9 KB | ||
| Others |  emd_41014_half_map_1.map.gz emd_41014_half_map_1.map.gz emd_41014_half_map_2.map.gz emd_41014_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41014 http://ftp.pdbj.org/pub/emdb/structures/EMD-41014 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41014 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41014 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41014.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41014.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DHA-FFA1-miniGq-Local | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: DHA-FFA1-miniGq-Local-half B
| File | emd_41014_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DHA-FFA1-miniGq-Local-half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
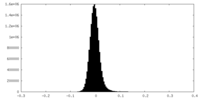
| Density Histograms |
-Half map: DHA-FFA1-miniGq-Local-half A
| File | emd_41014_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DHA-FFA1-miniGq-Local-half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
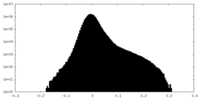
| Density Histograms |
- Sample components
Sample components
-Entire : DHA-FFA1-miniGq complex(mask on receptor)
| Entire | Name: DHA-FFA1-miniGq complex(mask on receptor) |
|---|---|
| Components |
|
-Supramolecule #1: DHA-FFA1-miniGq complex(mask on receptor)
| Supramolecule | Name: DHA-FFA1-miniGq complex(mask on receptor) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 305318 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X