+Search query
-Structure paper
| Title | A dimeric proteomimetic prevents SARS-CoV-2 infection by dimerizing the spike protein. |
|---|---|
| Journal, issue, pages | Nat Chem Biol, Vol. 18, Issue 10, Page 1046-1055, Year 2022 |
| Publish date | Jun 2, 2022 |
 Authors Authors | Bhavesh Khatri / Ishika Pramanick / Sameer Kumar Malladi / Raju S Rajmani / Sahil Kumar / Pritha Ghosh / Nayanika Sengupta / R Rahisuddin / Narender Kumar / S Kumaran / Rajesh P Ringe / Raghavan Varadarajan / Somnath Dutta / Jayanta Chatterjee /  |
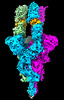
| PubMed Abstract | Protein tertiary structure mimetics are valuable tools to target large protein-protein interaction interfaces. Here, we demonstrate a strategy for designing dimeric helix-hairpin motifs from a ...Protein tertiary structure mimetics are valuable tools to target large protein-protein interaction interfaces. Here, we demonstrate a strategy for designing dimeric helix-hairpin motifs from a previously reported three-helix-bundle miniprotein that targets the receptor-binding domain (RBD) of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Through truncation of the third helix and optimization of the interhelical loop residues of the miniprotein, we developed a thermostable dimeric helix-hairpin. The dimeric four-helix bundle competes with the human angiotensin-converting enzyme 2 (ACE2) in binding to RBD with 2:2 stoichiometry. Cryogenic-electron microscopy revealed the formation of dimeric spike ectodomain trimer by the four-helix bundle, where all the three RBDs from either spike protein are attached head-to-head in an open conformation, revealing a novel mechanism for virus neutralization. The proteomimetic protects hamsters from high dose viral challenge with replicative SARS-CoV-2 viruses, demonstrating the promise of this class of peptides that inhibit protein-protein interaction through target dimerization. |
 External links External links |  Nat Chem Biol / Nat Chem Biol /  PubMed:35654847 / PubMed:35654847 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 4.47 - 5.41 Å |
| Structure data |  EMDB-32388: Cryo-EM 3D model of the 3-RBD up dimeric spike protein of SARS-CoV2 in the presence of SIH-5 EMDB-33042: Cryo-EM 3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5. |
| Chemicals |  ChemComp-NAG: |
| Source |
|
 Keywords Keywords |  VIRAL PROTEIN / VIRAL PROTEIN /  spike protein / spike protein /  SARS-CoV2 / SARS-CoV2 /  complex complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers