+Search query
-Structure paper
| Title | Fragment-based drug discovery using cryo-EM. |
|---|---|
| Journal, issue, pages | Drug Discov Today, Vol. 25, Issue 3, Page 485-490, Year 2020 |
| Publish date | Dec 23, 2019 |
 Authors Authors | Michael Saur / Michael J Hartshorn / Jing Dong / Judith Reeks / Gabor Bunkoczi / Harren Jhoti / Pamela A Williams /  |
| PubMed Abstract | Recent advances in electron cryo-microscopy (cryo-EM) structure determination have pushed the resolutions obtainable by the method into the range widely considered to be of utility for drug discovery. ...Recent advances in electron cryo-microscopy (cryo-EM) structure determination have pushed the resolutions obtainable by the method into the range widely considered to be of utility for drug discovery. Here, we review the use of cryo-EM in fragment-based drug discovery (FBDD) based on in-house method development. We demonstrate not only that cryo-EM can reveal details of the molecular interactions between fragments and a protein, but also that the current reproducibility, quality, and throughput are compatible with FBDD. We exemplify this using the test system β-galactosidase (Bgal) and the oncology target pyruvate kinase 2 (PKM2). |
 External links External links |  Drug Discov Today / Drug Discov Today /  PubMed:31877353 PubMed:31877353 |
| Methods | EM (single particle) |
| Resolution | 2.2 - 3.2 Å |
| Structure data | EMDB-10563, PDB-6tsh: EMDB-10564, PDB-6tsk: EMDB-10574, PDB-6tte: EMDB-10575, PDB-6ttf: EMDB-10576, PDB-6tth: EMDB-10577, PDB-6tti: EMDB-10584, PDB-6ttq: |
| Chemicals |  ChemComp-MG:  ChemComp-DGJ:  ChemComp-HOH: 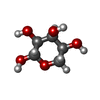 ChemComp-0MK:  ChemComp-PTQ: 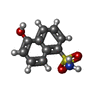 ChemComp-LZ2: 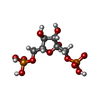 ChemComp-FBP: 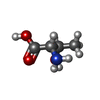 ChemComp-THR: 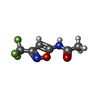 ChemComp-NXE:  ChemComp-DMS: 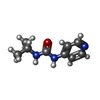 ChemComp-NXH: |
| Source |
|
 Keywords Keywords | SUGAR BINDING PROTEIN / Bgal / nojirimycin / L-ribose /  PETG / PETG /  CYTOSOLIC PROTEIN / CYTOSOLIC PROTEIN /  PKM2 / FBDD / L-threonine PKM2 / FBDD / L-threonine |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




















