+Search query
-Structure paper
| Title | Structural basis for variant-specific neuroligin-binding by α-neurexin. |
|---|---|
| Journal, issue, pages | PLoS One, Vol. 6, Issue 4, Page e19411, Year 2011 |
| Publish date | Apr 28, 2011 |
 Authors Authors | Hiroki Tanaka / Terukazu Nogi / Norihisa Yasui / Kenji Iwasaki / Junichi Takagi /  |
| PubMed Abstract | Neurexins (Nrxs) are presynaptic membrane proteins with a single membrane-spanning domain that mediate asymmetric trans-synaptic cell adhesion by binding to their postsynaptic receptor neuroligins. ...Neurexins (Nrxs) are presynaptic membrane proteins with a single membrane-spanning domain that mediate asymmetric trans-synaptic cell adhesion by binding to their postsynaptic receptor neuroligins. α-Nrx has a large extracellular region comprised of multiple copies of laminin, neurexin, sex-hormone-binding globulin (LNS) domains and epidermal growth factor (EGF) modules, while that of β-Nrx has but a single LNS domain. It has long been known that the larger α-Nrx and the shorter β-Nrx show distinct binding behaviors toward different isoforms/variants of neuroligins, although the underlying mechanism has yet to be elucidated. Here, we describe the crystal structure of a fragment corresponding to the C-terminal one-third of the Nrx1α ectodomain, consisting of LNS5-EGF3-LNS6. The 2.3 Å-resolution structure revealed the presence of a domain configuration that was rigidified by inter-domain contacts, as opposed to the more common flexible "beads-on-a-string" arrangement. Although the neuroligin-binding site on the LNS6 domain was completely exposed, the location of the α-Nrx specific LNS5-EGF3 segment proved incompatible with the loop segment inserted in the B+ neuroligin variant, which explains the variant-specific neuroligin recognition capability observed in α-Nrx. This, combined with a low-resolution molecular envelope obtained by a single particle reconstruction performed on negatively stained full-length Nrx1α sample, allowed us to derive a structural model of the α-Nrx ectodomain. This model will help us understand not only how the large α-Nrx ectodomain is accommodated in the synaptic cleft, but also how the trans-synaptic adhesion mediated by α- and β-Nrxs could differentially affect synaptic structure and function. |
 External links External links |  PLoS One / PLoS One /  PubMed:21552542 / PubMed:21552542 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.3 Å |
| Structure data | 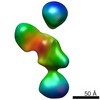 EMDB-5270:  PDB-3asi: |
| Chemicals |  ChemComp-CA:  ChemComp-NAG:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords |  CELL ADHESION / CELL ADHESION /  Beta-sandwich / synapse maturation / Beta-sandwich / synapse maturation /  Neuroligin / Neuroligin /  N-glycosylation / N-glycosylation /  Membrane Membrane |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




