+Search query
-Structure paper
| Title | SARS-COV-2 Omicron variants conformationally escape a rare quaternary antibody binding mode. |
|---|---|
| Journal, issue, pages | Commun Biol, Vol. 6, Issue 1, Page 1250, Year 2023 |
| Publish date | Dec 11, 2023 |
 Authors Authors | Jule Goike / Ching-Lin Hsieh / Andrew P Horton / Elizabeth C Gardner / Ling Zhou / Foteini Bartzoka / Nianshuang Wang / Kamyab Javanmardi / Andrew Herbert / Shawn Abbassi / Xuping Xie / Hongjie Xia / Pei-Yong Shi / Rebecca Renberg / Thomas H Segall-Shapiro / Cynthia I Terrace / Wesley Wu / Raghav Shroff / Michelle Byrom / Andrew D Ellington / Edward M Marcotte / James M Musser / Suresh V Kuchipudi / Vivek Kapur / George Georgiou / Scott C Weaver / John M Dye / Daniel R Boutz / Jason S McLellan / Jimmy D Gollihar /  |
| PubMed Abstract | The ongoing evolution of SARS-CoV-2 into more easily transmissible and infectious variants has provided unprecedented insight into mutations enabling immune escape. Understanding how these mutations ...The ongoing evolution of SARS-CoV-2 into more easily transmissible and infectious variants has provided unprecedented insight into mutations enabling immune escape. Understanding how these mutations affect the dynamics of antibody-antigen interactions is crucial to the development of broadly protective antibodies and vaccines. Here we report the characterization of a potent neutralizing antibody (N3-1) identified from a COVID-19 patient during the first disease wave. Cryogenic electron microscopy revealed a quaternary binding mode that enables direct interactions with all three receptor-binding domains of the spike protein trimer, resulting in extraordinary avidity and potent neutralization of all major variants of concern until the emergence of Omicron. Structure-based rational design of N3-1 mutants improved binding to all Omicron variants but only partially restored neutralization of the conformationally distinct Omicron BA.1. This study provides new insights into immune evasion through changes in spike protein dynamics and highlights considerations for future conformationally biased multivalent vaccine designs. |
 External links External links |  Commun Biol / Commun Biol /  PubMed:38082099 / PubMed:38082099 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.79 - 3.2 Å |
| Structure data | EMDB-41374, PDB-8tm1: EMDB-41382, PDB-8tma: 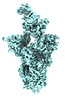 EMDB-41399: Antibody N3-1 bound to SARS-CoV-2 spike |
| Source |
|
 Keywords Keywords |  VIRAL PROTEIN/IMMUNE SYSTEM / SARS-CoV-2 spike / VIRAL PROTEIN/IMMUNE SYSTEM / SARS-CoV-2 spike /  neutralizing antibody / RBD-directed antibody / quaternary epitope / neutralizing antibody / RBD-directed antibody / quaternary epitope /  VIRAL PROTEIN / VIRAL PROTEIN /  VIRAL PROTEIN-IMMUNE SYSTEM complex VIRAL PROTEIN-IMMUNE SYSTEM complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers