+Search query
-Structure paper
| Title | Cdt1 stabilizes an open MCM ring for helicase loading. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 8, Page 15720, Year 2017 |
| Publish date | Jun 23, 2017 |
 Authors Authors | Jordi Frigola / Jun He / Kerstin Kinkelin / Valerie E Pye / Ludovic Renault / Max E Douglas / Dirk Remus / Peter Cherepanov / Alessandro Costa / John F X Diffley /   |
| PubMed Abstract | ORC, Cdc6 and Cdt1 act together to load hexameric MCM, the motor of the eukaryotic replicative helicase, into double hexamers at replication origins. Here we show that Cdt1 interacts with MCM ...ORC, Cdc6 and Cdt1 act together to load hexameric MCM, the motor of the eukaryotic replicative helicase, into double hexamers at replication origins. Here we show that Cdt1 interacts with MCM subunits Mcm2, 4 and 6, which both destabilizes the Mcm2-5 interface and inhibits MCM ATPase activity. Using X-ray crystallography, we show that Cdt1 contains two winged-helix domains in the C-terminal half of the protein and a catalytically inactive dioxygenase-related N-terminal domain, which is important for MCM loading, but not for subsequent replication. We used these structures together with single-particle electron microscopy to generate three-dimensional models of MCM complexes. These show that Cdt1 stabilizes MCM in a left-handed spiral open at the Mcm2-5 gate. We propose that Cdt1 acts as a brace, holding MCM open for DNA entry and bound to ATP until ORC-Cdc6 triggers ATP hydrolysis by MCM, promoting both Cdt1 ejection and MCM ring closure. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:28643783 / PubMed:28643783 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.8 - 20.4 Å |
| Structure data | 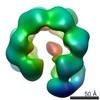 EMDB-3679: 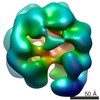 EMDB-3680: 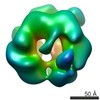 EMDB-3681:  PDB-5me9:  PDB-5mea:  PDB-5meb:  PDB-5mec: |
| Chemicals |  ChemComp-SO4:  ChemComp-GOL:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords |  CELL CYCLE / Cdt1 / MCM / CELL CYCLE / Cdt1 / MCM /  winged helix / winged helix /  yeast / yeast /  DNA replication DNA replication |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




