+Search query
-Structure paper
| Title | Architecture and activation of human muscle phosphorylase kinase. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 2719, Year 2024 |
| Publish date | Mar 28, 2024 |
 Authors Authors | Xiaoke Yang / Mingqi Zhu / Xue Lu / Yuxin Wang / Junyu Xiao /  |
| PubMed Abstract | The study of phosphorylase kinase (PhK)-regulated glycogen metabolism has contributed to the fundamental understanding of protein phosphorylation; however, the molecular mechanism of PhK remains ...The study of phosphorylase kinase (PhK)-regulated glycogen metabolism has contributed to the fundamental understanding of protein phosphorylation; however, the molecular mechanism of PhK remains poorly understood. Here we present the high-resolution cryo-electron microscopy structures of human muscle PhK. The 1.3-megadalton PhK αβγδ hexadecamer consists of a tetramer of tetramer, wherein four αβγδ modules are connected by the central β scaffold. The α- and β-subunits possess glucoamylase-like domains, but exhibit no detectable enzyme activities. The α-subunit serves as a bridge between the β-subunit and the γδ subcomplex, and facilitates the γ-subunit to adopt an autoinhibited state. Ca-free calmodulin (δ-subunit) binds to the γ-subunit in a compact conformation. Upon binding of Ca, a conformational change occurs, allowing for the de-inhibition of the γ-subunit through a spring-loaded mechanism. We also reveal an ADP-binding pocket in the β-subunit, which plays a role in allosterically enhancing PhK activity. These results provide molecular insights of this important kinase complex. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38548794 / PubMed:38548794 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.1 Å |
| Structure data | EMDB-36212, PDB-8jfk: EMDB-36213, PDB-8jfl: EMDB-36214: local map of hPhK alpha-beta-gamma-delta subcomplex in inactive state EMDB-36215: local map of hPhK gamma-delta subcomplex in inactive state EMDB-36216: local map of hPhK alpha-gamma subcomplex in active state |
| Chemicals | 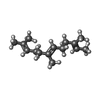 ChemComp-FAR:  ChemComp-ADP: |
| Source |
|
 Keywords Keywords |  CYTOSOLIC PROTEIN / glycogen phosphorylase b kinase / muscle isoform / inactive state / Ca2+ active state / hPhK alpha-gamma subcomplex / alpha-beta-gamma-delta subcomplex / gamma-delta subcomplex CYTOSOLIC PROTEIN / glycogen phosphorylase b kinase / muscle isoform / inactive state / Ca2+ active state / hPhK alpha-gamma subcomplex / alpha-beta-gamma-delta subcomplex / gamma-delta subcomplex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














