+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PhK holoenzyme in inactive state, muscle isoform | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycogen phosphorylase b kinase / muscle isoform / inactive state /  CYTOSOLIC PROTEIN CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information phosphorylase kinase / phosphorylase kinase /  phosphorylase kinase activity / phosphorylase kinase activity /  phosphorylase kinase complex / phosphorylase kinase complex /  tau-protein kinase / glycogen biosynthetic process / tau-protein kinase / glycogen biosynthetic process /  CaM pathway / Cam-PDE 1 activation / Sodium/Calcium exchangers / Calmodulin induced events / Reduction of cytosolic Ca++ levels ... CaM pathway / Cam-PDE 1 activation / Sodium/Calcium exchangers / Calmodulin induced events / Reduction of cytosolic Ca++ levels ... phosphorylase kinase / phosphorylase kinase /  phosphorylase kinase activity / phosphorylase kinase activity /  phosphorylase kinase complex / phosphorylase kinase complex /  tau-protein kinase / glycogen biosynthetic process / tau-protein kinase / glycogen biosynthetic process /  CaM pathway / Cam-PDE 1 activation / Sodium/Calcium exchangers / Calmodulin induced events / Reduction of cytosolic Ca++ levels / Activation of Ca-permeable Kainate Receptor / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / Loss of phosphorylation of MECP2 at T308 / CREB1 phosphorylation through the activation of Adenylate Cyclase / PKA activation / negative regulation of high voltage-gated calcium channel activity / CaMK IV-mediated phosphorylation of CREB / Glycogen breakdown (glycogenolysis) / negative regulation of calcium ion export across plasma membrane / organelle localization by membrane tethering / Activation of RAC1 downstream of NMDARs / regulation of cardiac muscle cell action potential / mitochondrion-endoplasmic reticulum membrane tethering / CLEC7A (Dectin-1) induces NFAT activation / autophagosome membrane docking / CaM pathway / Cam-PDE 1 activation / Sodium/Calcium exchangers / Calmodulin induced events / Reduction of cytosolic Ca++ levels / Activation of Ca-permeable Kainate Receptor / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / Loss of phosphorylation of MECP2 at T308 / CREB1 phosphorylation through the activation of Adenylate Cyclase / PKA activation / negative regulation of high voltage-gated calcium channel activity / CaMK IV-mediated phosphorylation of CREB / Glycogen breakdown (glycogenolysis) / negative regulation of calcium ion export across plasma membrane / organelle localization by membrane tethering / Activation of RAC1 downstream of NMDARs / regulation of cardiac muscle cell action potential / mitochondrion-endoplasmic reticulum membrane tethering / CLEC7A (Dectin-1) induces NFAT activation / autophagosome membrane docking /  tau-protein kinase activity / glycogen metabolic process / positive regulation of ryanodine-sensitive calcium-release channel activity / Negative regulation of NMDA receptor-mediated neuronal transmission / regulation of cell communication by electrical coupling involved in cardiac conduction / Unblocking of NMDA receptors, glutamate binding and activation / negative regulation of peptidyl-threonine phosphorylation / Synthesis of IP3 and IP4 in the cytosol / Phase 0 - rapid depolarisation / protein phosphatase activator activity / RHO GTPases activate PAKs / positive regulation of cyclic-nucleotide phosphodiesterase activity / positive regulation of phosphoprotein phosphatase activity / tau-protein kinase activity / glycogen metabolic process / positive regulation of ryanodine-sensitive calcium-release channel activity / Negative regulation of NMDA receptor-mediated neuronal transmission / regulation of cell communication by electrical coupling involved in cardiac conduction / Unblocking of NMDA receptors, glutamate binding and activation / negative regulation of peptidyl-threonine phosphorylation / Synthesis of IP3 and IP4 in the cytosol / Phase 0 - rapid depolarisation / protein phosphatase activator activity / RHO GTPases activate PAKs / positive regulation of cyclic-nucleotide phosphodiesterase activity / positive regulation of phosphoprotein phosphatase activity /  Long-term potentiation / Ion transport by P-type ATPases / Uptake and function of anthrax toxins / Calcineurin activates NFAT / Regulation of MECP2 expression and activity / Long-term potentiation / Ion transport by P-type ATPases / Uptake and function of anthrax toxins / Calcineurin activates NFAT / Regulation of MECP2 expression and activity /  catalytic complex / DARPP-32 events / detection of calcium ion / negative regulation of ryanodine-sensitive calcium-release channel activity / Smooth Muscle Contraction / RHO GTPases activate IQGAPs / regulation of cardiac muscle contraction / calcium channel inhibitor activity / cellular response to interferon-beta / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / catalytic complex / DARPP-32 events / detection of calcium ion / negative regulation of ryanodine-sensitive calcium-release channel activity / Smooth Muscle Contraction / RHO GTPases activate IQGAPs / regulation of cardiac muscle contraction / calcium channel inhibitor activity / cellular response to interferon-beta / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion /  Protein methylation / Protein methylation /  voltage-gated potassium channel complex / Activation of AMPK downstream of NMDARs / eNOS activation / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / regulation of calcium-mediated signaling / positive regulation of protein dephosphorylation / voltage-gated potassium channel complex / Activation of AMPK downstream of NMDARs / eNOS activation / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / regulation of calcium-mediated signaling / positive regulation of protein dephosphorylation /  titin binding / regulation of ryanodine-sensitive calcium-release channel activity / Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / Ion homeostasis / positive regulation of protein autophosphorylation / sperm midpiece / titin binding / regulation of ryanodine-sensitive calcium-release channel activity / Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / Ion homeostasis / positive regulation of protein autophosphorylation / sperm midpiece /  calcium channel complex / substantia nigra development / adenylate cyclase activator activity / Ras activation upon Ca2+ influx through NMDA receptor / calcium channel complex / substantia nigra development / adenylate cyclase activator activity / Ras activation upon Ca2+ influx through NMDA receptor /  regulation of heart rate / protein serine/threonine kinase activator activity / regulation of heart rate / protein serine/threonine kinase activator activity /  sarcomere / FCERI mediated Ca+2 mobilization / FCGR3A-mediated IL10 synthesis / VEGFR2 mediated vascular permeability / positive regulation of peptidyl-threonine phosphorylation / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / VEGFR2 mediated cell proliferation / sarcomere / FCERI mediated Ca+2 mobilization / FCGR3A-mediated IL10 synthesis / VEGFR2 mediated vascular permeability / positive regulation of peptidyl-threonine phosphorylation / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / VEGFR2 mediated cell proliferation /  regulation of cytokinesis / generation of precursor metabolites and energy / Translocation of SLC2A4 (GLUT4) to the plasma membrane / spindle microtubule / RAF activation / positive regulation of receptor signaling pathway via JAK-STAT / positive regulation of protein serine/threonine kinase activity / Transcriptional activation of mitochondrial biogenesis / Stimuli-sensing channels / regulation of cytokinesis / generation of precursor metabolites and energy / Translocation of SLC2A4 (GLUT4) to the plasma membrane / spindle microtubule / RAF activation / positive regulation of receptor signaling pathway via JAK-STAT / positive regulation of protein serine/threonine kinase activity / Transcriptional activation of mitochondrial biogenesis / Stimuli-sensing channels /  spindle pole / cellular response to type II interferon / response to calcium ion / RAS processing / calcium-dependent protein binding / Inactivation, recovery and regulation of the phototransduction cascade / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / G2/M transition of mitotic cell cycle / Signaling by BRAF and RAF1 fusions spindle pole / cellular response to type II interferon / response to calcium ion / RAS processing / calcium-dependent protein binding / Inactivation, recovery and regulation of the phototransduction cascade / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / G2/M transition of mitotic cell cycle / Signaling by BRAF and RAF1 fusionsSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Yang XK / Xiao JY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Architecture and activation of human muscle phosphorylase kinase. Authors: Xiaoke Yang / Mingqi Zhu / Xue Lu / Yuxin Wang / Junyu Xiao /  Abstract: The study of phosphorylase kinase (PhK)-regulated glycogen metabolism has contributed to the fundamental understanding of protein phosphorylation; however, the molecular mechanism of PhK remains ...The study of phosphorylase kinase (PhK)-regulated glycogen metabolism has contributed to the fundamental understanding of protein phosphorylation; however, the molecular mechanism of PhK remains poorly understood. Here we present the high-resolution cryo-electron microscopy structures of human muscle PhK. The 1.3-megadalton PhK αβγδ hexadecamer consists of a tetramer of tetramer, wherein four αβγδ modules are connected by the central β scaffold. The α- and β-subunits possess glucoamylase-like domains, but exhibit no detectable enzyme activities. The α-subunit serves as a bridge between the β-subunit and the γδ subcomplex, and facilitates the γ-subunit to adopt an autoinhibited state. Ca-free calmodulin (δ-subunit) binds to the γ-subunit in a compact conformation. Upon binding of Ca, a conformational change occurs, allowing for the de-inhibition of the γ-subunit through a spring-loaded mechanism. We also reveal an ADP-binding pocket in the β-subunit, which plays a role in allosterically enhancing PhK activity. These results provide molecular insights of this important kinase complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36212.map.gz emd_36212.map.gz | 276.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36212-v30.xml emd-36212-v30.xml emd-36212.xml emd-36212.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36212.png emd_36212.png | 37.5 KB | ||
| Filedesc metadata |  emd-36212.cif.gz emd-36212.cif.gz | 7.1 KB | ||
| Others |  emd_36212_half_map_1.map.gz emd_36212_half_map_1.map.gz emd_36212_half_map_2.map.gz emd_36212_half_map_2.map.gz | 301.4 MB 301.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36212 http://ftp.pdbj.org/pub/emdb/structures/EMD-36212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36212 | HTTPS FTP |
-Related structure data
| Related structure data |  8jfkMC  8jflC  8xy7C  8xyaC  8xybC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36212.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36212.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36212_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
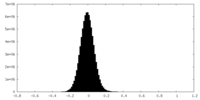
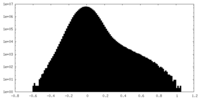
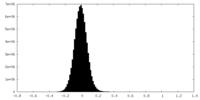
| Density Histograms |
-Half map: #1
| File | emd_36212_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : phosphorylase b kinase, muscle isoform
| Entire | Name: phosphorylase b kinase, muscle isoform |
|---|---|
| Components |
|
-Supramolecule #1: phosphorylase b kinase, muscle isoform
| Supramolecule | Name: phosphorylase b kinase, muscle isoform / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2, #4, #3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphorylase b kinase regulatory subunit beta
| Macromolecule | Name: Phosphorylase b kinase regulatory subunit beta / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 125.032961 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGAAGLTAE VSWKVLERRA RTKRSGSVYE PLKSINLPRP DNETLWDKLD HYYRIVKSTL LLYQSPTTGL FPTKTCGGDQ KAKIQDSLY CAAGAWALAL AYRRIDDDKG RTHELEHSAI KCMRGILYCY MRQADKVQQF KQDPRPTTCL HSVFNVHTGD E LLSYEEYG ...String: MAGAAGLTAE VSWKVLERRA RTKRSGSVYE PLKSINLPRP DNETLWDKLD HYYRIVKSTL LLYQSPTTGL FPTKTCGGDQ KAKIQDSLY CAAGAWALAL AYRRIDDDKG RTHELEHSAI KCMRGILYCY MRQADKVQQF KQDPRPTTCL HSVFNVHTGD E LLSYEEYG HLQINAVSLY LLYLVEMISS GLQIIYNTDE VSFIQNLVFC VERVYRVPDF GVWERGSKYN NGSTELHSSS VG LAKAALE AINGFNLFGN QGCSWSVIFV DLDAHNRNRQ TLCSLLPRES RSHNTDAALL PCISYPAFAL DDEVLFSQTL DKV VRKLKG KYGFKRFLRD GYRTSLEDPN RCYYKPAEIK LFDGIECEFP IFFLYMMIDG VFRGNPKQVQ EYQDLLTPVL HHTT EGYPV VPKYYYVPAD FVEYEKNNPG SQKRFPSNCG RDGKLFLWGQ ALYIIAKLLA DELISPKDID PVQRYVPLKD QRNVS MRFS NQGPLENDLV VHVALIAESQ RLQVFLNTYG IQTQTPQQVE PIQIWPQQEL VKAYLQLGIN EKLGLSGRPD RPIGCL GTS KIYRILGKTV VCYPIIFDLS DFYMSQDVFL LIDDIKNALQ FIKQYWKMHG RPLFLVLIRE DNIRGSRFNP ILDMLAA LK KGIIGGVKVH VDRLQTLISG AVVEQLDFLR ISDTEELPEF KSFEELEPPK HSKVKRQSST PSAPELGQQP DVNISEWK D KPTHEILQKL NDCSCLASQA ILLGILLKRE GPNFITKEGT VSDHIERVYR RAGSQKLWLA VRYGAAFTQK FSSSIAPHI TTFLVHGKQV TLGAFGHEEE VISNPLSPRV IQNIIYYKCN THDEREAVIQ QELVIHIGWI ISNNPELFSG MLKIRIGWII HAMEYELQI RGGDKPALDL YQLSPSEVKQ LLLDILQPQQ NGRCWLNRRQ IDGSLNRTPT GFYDRVWQIL ERTPNGIIVA G KHLPQQPT LSDMTMYEMN FSLLVEDTLG NIDQPQYRQI VVELLMVVSI VLERNPELEF QDKVDLDRLV KEAFNEFQKD QS RLKEIEK QDDMTSFYNT PPLGKRGTCS YLTKAVMNLL LEGEVKPNND DPCLIS UniProtKB: Phosphorylase b kinase regulatory subunit beta |
-Macromolecule #2: Phosphorylase b kinase regulatory subunit alpha, skeletal muscle ...
| Macromolecule | Name: Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137.469422 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRSRSNSGVR LDGYARLVQQ TILCHQNPVT GLLPASYDQK DAWVRDNVYS ILAVWGLGLA YRKNADRDED KAKAYELEQS VVKLMRGLL HCMIRQVDKV ESFKYSQSTK DSLHAKYNTK TCATVVGDDQ WGHLQLDATS VYLLFLAQMT ASGLHIIHSL D EVNFIQNL ...String: MRSRSNSGVR LDGYARLVQQ TILCHQNPVT GLLPASYDQK DAWVRDNVYS ILAVWGLGLA YRKNADRDED KAKAYELEQS VVKLMRGLL HCMIRQVDKV ESFKYSQSTK DSLHAKYNTK TCATVVGDDQ WGHLQLDATS VYLLFLAQMT ASGLHIIHSL D EVNFIQNL VFYIEAAYKT ADFGIWERGD KTNQGISELN ASSVGMAKAA LEALDELDLF GVKGGPQSVI HVLADEVQHC QS ILNSLLP RASTSKEVDA SLLSVVSFPA FAVEDSQLVE LTKQEIITKL QGRYGCCRFL RDGYKTPKED PNRLYYEPAE LKL FENIEC EWPLFWTYFI LDGVFSGNAE QVQEYKEALE AVLIKGKNGV PLLPELYSVP PDRVDEEYQN PHTVDRVPMG KLPH MWGQS LYILGSLMAE GFLAPGEIDP LNRRFSTVPK PDVVVQVSIL AETEEIKTIL KDKGIYVETI AEVYPIRVQP ARILS HIYS SLGCNNRMKL SGRPYRHMGV LGTSKLYDIR KTIFTFTPQF IDQQQFYLAL DNKMIVEMLR TDLSYLCSRW RMTGQP TIT FPISHSMLDE DGTSLNSSIL AALRKMQDGY FGGARVQTGK LSEFLTTSCC THLSFMDPGP EGKLYSEDYD DNYDYLE SG NWMNDYDSTS HARCGDEVAR YLDHLLAHTA PHPKLAPTSQ KGGLDRFQAA VQTTCDLMSL VTKAKELHVQ NVHMYLPT K LFQASRPSFN LLDSPHPRQE NQVPSVRVEI HLPRDQSGEV DFKALVLQLK ETSSLQEQAD ILYMLYTMKG PDWNTELYN ERSATVRELL TELYGKVGEI RHWGLIRYIS GILRKKVEAL DEACTDLLSH QKHLTVGLPP EPREKTISAP LPYEALTQLI DEASEGDMS ISILTQEIMV YLAMYMRTQP GLFAEMFRLR IGLIIQVMAT ELAHSLRCSA EEATEGLMNL SPSAMKNLLH H ILSGKEFG VERSVRPTDS NVSPAISIHE IGAVGATKTE RTGIMQLKSE IKQVEFRRLS ISAESQSPGT SMTPSSGSFP SA YDQQSSK DSRQGQWQRR RRLDGALNRV PVGFYQKVWK VLQKCHGLSV EGFVLPSSTT REMTPGEIKF SVHVESVLNR VPQ PEYRQL LVEAILVLTM LADIEIHSIG SIIAVEKIVH IANDLFLQEQ KTLGADDTML AKDPASGICT LLYDSAPSGR FGTM TYLSK AAATYVQEFL PHSICAMQ UniProtKB: Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform |
-Macromolecule #3: Calmodulin-1
| Macromolecule | Name: Calmodulin-1 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.852545 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADQLTEEQI AEFKEAFSLF DKDGDGTITT KELGTVMRSL GQNPTEAELQ DMINEVDADG NGTIDFPEFL TMMARKMKDT DSEEEIREA FRVFDKDGNG YISAAELRHV MTNLGEKLTD EEVDEMIREA DIDGDGQVNY EEFVQMMTAK UniProtKB:  Calmodulin-1 Calmodulin-1 |
-Macromolecule #4: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/hea...
| Macromolecule | Name: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO / EC number:  phosphorylase kinase phosphorylase kinase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.084672 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTRDEALPDS HSAQDFYENY EPKEILGRGV SSVVRRCIHK PTSQEYAVKV IDVTGGGSFS PEEVRELREA TLKEVDILRK VSGHPNIIQ LKDTYETNTF FFLVFDLMKR GELFDYLTEK VTLSEKETRK IMRALLEVIC TLHKLNIVHR DLKPENILLD D NMNIKLTD ...String: MTRDEALPDS HSAQDFYENY EPKEILGRGV SSVVRRCIHK PTSQEYAVKV IDVTGGGSFS PEEVRELREA TLKEVDILRK VSGHPNIIQ LKDTYETNTF FFLVFDLMKR GELFDYLTEK VTLSEKETRK IMRALLEVIC TLHKLNIVHR DLKPENILLD D NMNIKLTD FGFSCQLEPG ERLREVCGTP SYLAPEIIEC SMNEDHPGYG KEVDMWSTGV IMYTLLAGSP PFWHRKQMLM LR MIMSGNY QFGSPEWDDY SDTVKDLVSR FLVVQPQNRY TAEEALAHPF FQQYLVEEVR HFSPRGKFKV IALTVLASVR IYY QYRRVK PVTREIVIRD PYALRPLRRL IDAYAFRIYG HWVKKGQQQN RAALFENTPK AVLLSLAEED Y UniProtKB: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform |
-Macromolecule #5: FARNESYL
| Macromolecule | Name: FARNESYL / type: ligand / ID: 5 / Number of copies: 8 / Formula: FAR |
|---|---|
| Molecular weight | Theoretical: 206.367 Da |
| Chemical component information |  ChemComp-FAR: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.1 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 623973 |
 Movie
Movie Controller
Controller






























 Z
Z Y
Y X
X