+Search query
-Structure paper
| Title | Cms1 coordinates stepwise local 90S pre-ribosome assembly with timely snR83 release. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 41, Issue 8, Page 111684, Year 2022 |
| Publish date | Nov 22, 2022 |
 Authors Authors | Benjamin Lau / Olga Beine-Golovchuk / Markus Kornprobst / Jingdong Cheng / Dieter Kressler / Beáta Jády / Tamás Kiss / Roland Beckmann / Ed Hurt /     |
| PubMed Abstract | Ribosome synthesis begins in the nucleolus with 90S pre-ribosome construction, but little is known about how the many different snoRNAs that modify the pre-rRNA are timely guided to their target ...Ribosome synthesis begins in the nucleolus with 90S pre-ribosome construction, but little is known about how the many different snoRNAs that modify the pre-rRNA are timely guided to their target sites. Here, we report a role for Cms1 in such a process. Initially, we discovered CMS1 as a null suppressor of a nop14 mutant impaired in Rrp12-Enp1 factor recruitment to the 90S. Further investigations detected Cms1 at the 18S rRNA 3' major domain of an early 90S that carried H/ACA snR83, which is known to guide pseudouridylation at two target sites within the same subdomain. Cms1 co-precipitates with many 90S factors, but Rrp12-Enp1 encircling the 3' major domain in the mature 90S is decreased. We suggest that Cms1 associates with the 3' major domain during early 90S biogenesis to restrict premature Rrp12-Enp1 binding but allows snR83 to timely perform its modification role before the next 90S assembly steps coupled with Cms1 release take place. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:36417864 / PubMed:36417864 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.7 - 6.7 Å |
| Structure data |  EMDB-34283: Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae (Nop14-5Ala), state post-A1 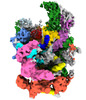 EMDB-34284: Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae (Rcl1/Cms1 KO)  EMDB-34285: Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae (Nop14 5Ala, Cms1 KO), state B2  EMDB-34286: Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae (Nop14 5Ala, Cms1 KO), state b |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Saccaromyces cerevisiae (brewer's yeast)
Saccaromyces cerevisiae (brewer's yeast)