[English] 日本語
 Yorodumi
Yorodumi- EMDB-34286: Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cere... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
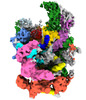
| Title | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae (Nop14 5Ala, Cms1 KO), state b | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Cheng J / Lau B / Hurt E / Beckmann R | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Cms1 coordinates stepwise local 90S pre-ribosome assembly with timely snR83 release. Authors: Benjamin Lau / Olga Beine-Golovchuk / Markus Kornprobst / Jingdong Cheng / Dieter Kressler / Beáta Jády / Tamás Kiss / Roland Beckmann / Ed Hurt /     Abstract: Ribosome synthesis begins in the nucleolus with 90S pre-ribosome construction, but little is known about how the many different snoRNAs that modify the pre-rRNA are timely guided to their target ...Ribosome synthesis begins in the nucleolus with 90S pre-ribosome construction, but little is known about how the many different snoRNAs that modify the pre-rRNA are timely guided to their target sites. Here, we report a role for Cms1 in such a process. Initially, we discovered CMS1 as a null suppressor of a nop14 mutant impaired in Rrp12-Enp1 factor recruitment to the 90S. Further investigations detected Cms1 at the 18S rRNA 3' major domain of an early 90S that carried H/ACA snR83, which is known to guide pseudouridylation at two target sites within the same subdomain. Cms1 co-precipitates with many 90S factors, but Rrp12-Enp1 encircling the 3' major domain in the mature 90S is decreased. We suggest that Cms1 associates with the 3' major domain during early 90S biogenesis to restrict premature Rrp12-Enp1 binding but allows snR83 to timely perform its modification role before the next 90S assembly steps coupled with Cms1 release take place. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34286.map.gz emd_34286.map.gz | 244.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34286-v30.xml emd-34286-v30.xml emd-34286.xml emd-34286.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34286_fsc.xml emd_34286_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_34286.png emd_34286.png | 134 KB | ||
| Others |  emd_34286_half_map_1.map.gz emd_34286_half_map_1.map.gz emd_34286_half_map_2.map.gz emd_34286_half_map_2.map.gz | 338.2 MB 338 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34286 http://ftp.pdbj.org/pub/emdb/structures/EMD-34286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34286 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34286.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34286.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34286_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34286_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 90S pre-ribosome
| Entire | Name: 90S pre-ribosome |
|---|---|
| Components |
|
-Supramolecule #1: 90S pre-ribosome
| Supramolecule | Name: 90S pre-ribosome / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X