+Search query
-Structure paper
| Title | FAP106 is an interaction hub for assembling microtubule inner proteins at the cilium inner junction. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 5225, Year 2023 |
| Publish date | Aug 26, 2023 |
 Authors Authors | Michelle M Shimogawa / Angeline S Wijono / Hui Wang / Jiayan Zhang / Jihui Sha / Natasha Szombathy / Sabeeca Vadakkan / Paula Pelayo / Keya Jonnalagadda / James Wohlschlegel / Z Hong Zhou / Kent L Hill /  |
| PubMed Abstract | Motility of pathogenic protozoa depends on flagella (synonymous with cilia) with axonemes containing nine doublet microtubules (DMTs) and two singlet microtubules. Microtubule inner proteins (MIPs) ...Motility of pathogenic protozoa depends on flagella (synonymous with cilia) with axonemes containing nine doublet microtubules (DMTs) and two singlet microtubules. Microtubule inner proteins (MIPs) within DMTs influence axoneme stability and motility and provide lineage-specific adaptations, but individual MIP functions and assembly mechanisms are mostly unknown. Here, we show in the sleeping sickness parasite Trypanosoma brucei, that FAP106, a conserved MIP at the DMT inner junction, is required for trypanosome motility and functions as a critical interaction hub, directing assembly of several conserved and lineage-specific MIPs. We use comparative cryogenic electron tomography (cryoET) and quantitative proteomics to identify MIP candidates. Using RNAi knockdown together with fitting of AlphaFold models into cryoET maps, we demonstrate that one of these candidates, MC8, is a trypanosome-specific MIP required for parasite motility. Our work advances understanding of MIP assembly mechanisms and identifies lineage-specific motility proteins that are attractive targets to consider for therapeutic intervention. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37633952 / PubMed:37633952 /  PubMed Central PubMed Central |
| Methods | EM (subtomogram averaging) |
| Resolution | 16.5 - 24.0 Å |
| Structure data | 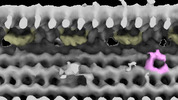 EMDB-28802: Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei FAP106 knockdown mutant 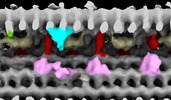 EMDB-28803: Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype  EMDB-28804: Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei MC8 Knockdown mutant 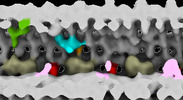 EMDB-28805: Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype Control 2 |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Trypanosoma brucei (eukaryote)
Trypanosoma brucei (eukaryote)