[English] 日本語
 Yorodumi
Yorodumi- EMDB-28803: Subtomographic average of flagellar axoneme doublet from Trypanos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 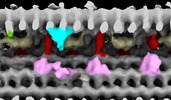 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
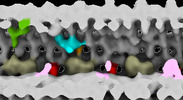
| Title | Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei Wildtype | |||||||||
 Map data Map data | Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei WildType Contol 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  flagella / flagella /  axoneme / MIPs / control data / axoneme / MIPs / control data /  STRUCTURAL PROTEIN STRUCTURAL PROTEIN | |||||||||
| Biological species |   Trypanosoma brucei (eukaryote) Trypanosoma brucei (eukaryote) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 18.5 Å cryo EM / Resolution: 18.5 Å | |||||||||
 Authors Authors | Shimogawa MM / Wang H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: FAP106 is an interaction hub for assembling microtubule inner proteins at the cilium inner junction. Authors: Michelle M Shimogawa / Angeline S Wijono / Hui Wang / Jiayan Zhang / Jihui Sha / Natasha Szombathy / Sabeeca Vadakkan / Paula Pelayo / Keya Jonnalagadda / James Wohlschlegel / Z Hong Zhou / Kent L Hill /  Abstract: Motility of pathogenic protozoa depends on flagella (synonymous with cilia) with axonemes containing nine doublet microtubules (DMTs) and two singlet microtubules. Microtubule inner proteins (MIPs) ...Motility of pathogenic protozoa depends on flagella (synonymous with cilia) with axonemes containing nine doublet microtubules (DMTs) and two singlet microtubules. Microtubule inner proteins (MIPs) within DMTs influence axoneme stability and motility and provide lineage-specific adaptations, but individual MIP functions and assembly mechanisms are mostly unknown. Here, we show in the sleeping sickness parasite Trypanosoma brucei, that FAP106, a conserved MIP at the DMT inner junction, is required for trypanosome motility and functions as a critical interaction hub, directing assembly of several conserved and lineage-specific MIPs. We use comparative cryogenic electron tomography (cryoET) and quantitative proteomics to identify MIP candidates. Using RNAi knockdown together with fitting of AlphaFold models into cryoET maps, we demonstrate that one of these candidates, MC8, is a trypanosome-specific MIP required for parasite motility. Our work advances understanding of MIP assembly mechanisms and identifies lineage-specific motility proteins that are attractive targets to consider for therapeutic intervention. | |||||||||
| History |
|
- Structure visualization
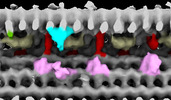
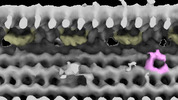
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28803.map.gz emd_28803.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28803-v30.xml emd-28803-v30.xml emd-28803.xml emd-28803.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28803.png emd_28803.png | 88.3 KB | ||
| Others |  emd_28803_half_map_1.map.gz emd_28803_half_map_1.map.gz emd_28803_half_map_2.map.gz emd_28803_half_map_2.map.gz | 3.4 MB 3.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28803 http://ftp.pdbj.org/pub/emdb/structures/EMD-28803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28803 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28803.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28803.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei WildType Contol 1 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 6.759 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Subtomographic average of flagellar axoneme doublet from Trypanosoma...
| File | emd_28803_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei WildType Contol 1 (half map 1) | ||||||||||||
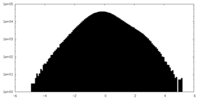
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Subtomographic average of flagellar axoneme doublet from Trypanosoma...
| File | emd_28803_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomographic average of flagellar axoneme doublet from Trypanosoma brucei WildType Contol 1 (half map 2) | ||||||||||||
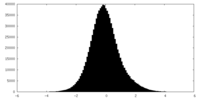
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Detergent-extracted flagellum of Trypanosoma brucei
| Entire | Name: Detergent-extracted flagellum of Trypanosoma brucei |
|---|---|
| Components |
|
-Supramolecule #1: Detergent-extracted flagellum of Trypanosoma brucei
| Supramolecule | Name: Detergent-extracted flagellum of Trypanosoma brucei / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei (eukaryote) / Organelle: flagella Trypanosoma brucei (eukaryote) / Organelle: flagella |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 2.68 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Extraction | Number tomograms: 20 / Number images used: 1616 |
|---|---|
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: PEET |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 18.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: PEET / Details: The FSC was performed by calcUnbiasedFSC from PEET / Number subtomograms used: 1014 |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X