+Search query
-Structure paper
| Title | Cryo EM structures map a post vaccination polyclonal antibody response to canine parvovirus. |
|---|---|
| Journal, issue, pages | Commun Biol, Vol. 6, Issue 1, Page 955, Year 2023 |
| Publish date | Sep 19, 2023 |
 Authors Authors | Samantha R Hartmann / Andrew J Charnesky / Simon P Früh / Robert A López-Astacio / Wendy S Weichert / Nadia DiNunno / Sung Hung Cho / Carol M Bator / Colin R Parrish / Susan L Hafenstein /  |
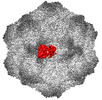
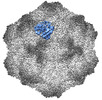
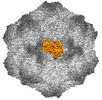
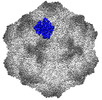
| PubMed Abstract | Canine parvovirus (CPV) is an important pathogen that emerged by cross-species transmission to cause severe disease in dogs. To understand the host immune response to vaccination, sera from dogs ...Canine parvovirus (CPV) is an important pathogen that emerged by cross-species transmission to cause severe disease in dogs. To understand the host immune response to vaccination, sera from dogs immunized with parvovirus are obtained, the polyclonal antibodies are purified and used to solve the high resolution cryo EM structures of the polyclonal Fab-virus complexes. We use a custom software, Icosahedral Subparticle Extraction and Correlated Classification (ISECC) to perform subparticle analysis and reconstruct polyclonal Fab-virus complexes from two different dogs eight and twelve weeks post vaccination. In the resulting polyclonal Fab-virus complexes there are a total of five distinct Fabs identified. In both cases, any of the five antibodies identified would interfere with receptor binding. This polyclonal mapping approach identifies a specific, limited immune response to the live vaccine virus and allows us to investigate the binding of multiple different antibodies or ligands to virus capsids. |
 External links External links |  Commun Biol / Commun Biol /  PubMed:37726539 / PubMed:37726539 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.0 - 3.8 Å |
| Structure data | EMDB-26786, PDB-7utp: EMDB-26787, PDB-7utr: EMDB-26788, PDB-7uts: EMDB-26789, PDB-7utu: EMDB-26790, PDB-7utv: |
| Source |
|
 Keywords Keywords | VIRUS/IMMUNE SYSTEM / CPV / polyclonal Fab /  A site / A site /  vaccination / vaccination /  VIRUS / VIRUS-IMMUNE SYSTEM complex / B site VIRUS / VIRUS-IMMUNE SYSTEM complex / B site |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers