+Search query
-Structure paper
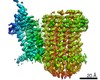
| Title | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. |
|---|---|
| Journal, issue, pages | Nature, Vol. 589, Issue 7840, Page 143-147, Year 2021 |
| Publish date | Dec 9, 2020 |
 Authors Authors | Hui Guo / Gautier M Courbon / Stephanie A Bueler / Juntao Mai / Jun Liu / John L Rubinstein /  |
| PubMed Abstract | Tuberculosis-the world's leading cause of death by infectious disease-is increasingly resistant to current first-line antibiotics. The bacterium Mycobacterium tuberculosis (which causes tuberculosis) ...Tuberculosis-the world's leading cause of death by infectious disease-is increasingly resistant to current first-line antibiotics. The bacterium Mycobacterium tuberculosis (which causes tuberculosis) can survive low-energy conditions, allowing infections to remain dormant and decreasing their susceptibility to many antibiotics. Bedaquiline was developed in 2005 from a lead compound identified in a phenotypic screen against Mycobacterium smegmatis. This drug can sterilize even latent M. tuberculosis infections and has become a cornerstone of treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Bedaquiline targets the mycobacterial ATP synthase, which is an essential enzyme in the obligate aerobic Mycobacterium genus, but how it binds the intact enzyme is unknown. Here we determined cryo-electron microscopy structures of M. smegmatis ATP synthase alone and in complex with bedaquiline. The drug-free structure suggests that hook-like extensions from the α-subunits prevent the enzyme from running in reverse, inhibiting ATP hydrolysis and preserving energy in hypoxic conditions. Bedaquiline binding induces large conformational changes in the ATP synthase, creating tight binding pockets at the interface of subunits a and c that explain the potency of this drug as an antibiotic for tuberculosis. |
 External links External links |  Nature / Nature /  PubMed:33299175 PubMed:33299175 |
| Methods | EM (single particle) |
| Resolution | 3.2 - 3.7 Å |
| Structure data | EMDB-22311, PDB-7jg5: EMDB-22312: Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 EMDB-22313: Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 3 EMDB-22314: Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 EMDB-22315: Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 2 EMDB-22316, PDB-7jga:  EMDB-22317:  EMDB-22318:  EMDB-22319: EMDB-22320, PDB-7jgb: EMDB-22321, PDB-7jgc:  EMDB-22322: |
| Chemicals |  ChemComp-ATP:  ChemComp-MG:  ChemComp-PO4:  ChemComp-ADP: 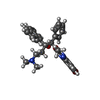 ChemComp-BQ1: |
| Source |
|
 Keywords Keywords |  TRANSLOCASE / TRANSLOCASE /  Bedaquiline / Bedaquiline /  Sirturo / Sirturo /  TMC207 / T207910 / TMC207 / T207910 /  ATP synthase / ATP synthase /  mycobacteria / mycobacteria /  tuberculosis / tuberculosis /  HYDROLASE HYDROLASE |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers