+Search query
-Structure paper
| Title | Ribosomal stalk-captured CARF-RelE ribonuclease inhibits translation following CRISPR signaling. |
|---|---|
| Journal, issue, pages | Science, Vol. 382, Issue 6674, Page 1036-1041, Year 2023 |
| Publish date | Nov 30, 2023 |
 Authors Authors | Irmantas Mogila / Giedre Tamulaitiene / Konstanty Keda / Albertas Timinskas / Audrone Ruksenaite / Giedrius Sasnauskas / Česlovas Venclovas / Virginijus Siksnys / Gintautas Tamulaitis |
| PubMed Abstract | Prokaryotic type III CRISPR-Cas antiviral systems employ cyclic oligoadenylate (cA) signaling to activate a diverse range of auxiliary proteins that reinforce the CRISPR-Cas defense. Here we ...Prokaryotic type III CRISPR-Cas antiviral systems employ cyclic oligoadenylate (cA) signaling to activate a diverse range of auxiliary proteins that reinforce the CRISPR-Cas defense. Here we characterize a class of cA-dependent effector proteins named CRISPR-Cas-associated messenger RNA (mRNA) interferase 1 (Cami1) consisting of a CRISPR-associated Rossmann fold sensor domain fused to winged helix-turn-helix and a RelE-family mRNA interferase domain. Upon activation by cyclic tetra-adenylate (cA), Cami1 cleaves mRNA exposed at the ribosomal A-site thereby depleting mRNA and leading to cell growth arrest. The structures of apo-Cami1 and the ribosome-bound Cami1-cA complex delineate the conformational changes that lead to Cami1 activation and the mechanism of Cami1 binding to a bacterial ribosome, revealing unexpected parallels with eukaryotic ribosome-inactivating proteins. |
 External links External links |  Science / Science /  PubMed:38033086 PubMed:38033086 |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.7 - 3.67 Å |
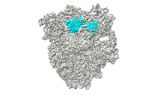
| Structure data | 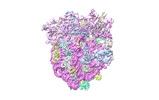 EMDB-17663: Cami1-ribosome cryoEM-map 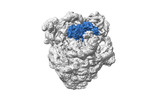 EMDB-17664: Cami1-ribosome local refinement map with mask on Cami1 and L12-Cter 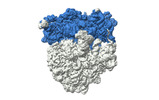 EMDB-17665: Cami1-ribosome local refinement map with mask on 30S subunit  EMDB-17666: Cami1-ribosome local refinement map with mask on 50S subunit EMDB-17667, PDB-8phj:  PDB-8phb: |
| Chemicals |  ChemComp-HOH:  ChemComp-ZN:  ChemComp-MG: |
| Source |
|
 Keywords Keywords |  HYDROLASE / HYDROLASE /  CRISPR / cyclic oligoadenylate / CRISPR / cyclic oligoadenylate /  RNAse / RelE-toxin / RNAse / RelE-toxin /  RIBOSOME RIBOSOME |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers