+Search query
-Structure paper
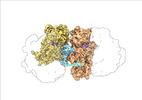
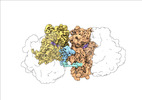
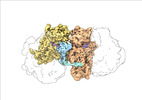
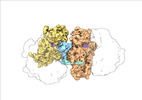
| Title | Transient disome complex formation in native polysomes during ongoing protein synthesis captured by cryo-EM. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 1756, Year 2024 |
| Publish date | Feb 26, 2024 |
 Authors Authors | Timo Flügel / Magdalena Schacherl / Anett Unbehaun / Birgit Schroeer / Marylena Dabrowski / Jörg Bürger / Thorsten Mielke / Thiemo Sprink / Christoph A Diebolder / Yollete V Guillén Schlippe / Christian M T Spahn /  |
| PubMed Abstract | Structural studies of translating ribosomes traditionally rely on in vitro assembly and stalling of ribosomes in defined states. To comprehensively visualize bacterial translation, we reactivated ex ...Structural studies of translating ribosomes traditionally rely on in vitro assembly and stalling of ribosomes in defined states. To comprehensively visualize bacterial translation, we reactivated ex vivo-derived E. coli polysomes in the PURE in vitro translation system and analyzed the actively elongating polysomes by cryo-EM. We find that 31% of 70S ribosomes assemble into disome complexes that represent eight distinct functional states including decoding and termination intermediates, and a pre-nucleophilic attack state. The functional diversity of disome complexes together with RNase digest experiments suggests that paused disome complexes transiently form during ongoing elongation. Structural analysis revealed five disome interfaces between leading and queueing ribosomes that undergo rearrangements as the leading ribosome traverses through the elongation cycle. Our findings reveal at the molecular level how bL9's CTD obstructs the factor binding site of queueing ribosomes to thwart harmful collisions and illustrate how translation dynamics reshape inter-ribosomal contacts. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38409277 / PubMed:38409277 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.09 - 9.11 Å |
| Structure data |  EMDB-17274: Escherichia coli paused disome complex (decoding | rotated PRE)  EMDB-17275: Escherichia coli paused disome complex (decoding | closed non-rot. PRE)  EMDB-17276: Escherichia coli paused disome complex (decoding | POST)  EMDB-17277: Escherichia coli paused disome complex (partially accommodated A-tRNA | closed non-rotated PRE)  EMDB-17278: Escherichia coli paused disome complex (partially accommodated A-tRNA | rotated PRE)  EMDB-17279: Escherichia coli paused disome complex (open non-rotated PRE | closed non-rotated PRE)  EMDB-17280: Escherichia coli paused disome complex (open non-rotated PRE | closed non-rotated PRE)  EMDB-17281: Escherichia coli paused disome complex (open non-rotated PRE | rotated PRE)  EMDB-17282: Escherichia coli paused disome complex (open non-rotated PRE | POST)  EMDB-17283: Escherichia coli paused disome complex (closed non-rotated PRE | closed non-rotated PRE)  EMDB-17284: Escherichia coli paused disome complex (closed non-rotated PRE | rotated PRE)  EMDB-17285: Escherichia coli paused disome complex (closed non-rotated PRE | POST)  EMDB-17286: Escherichia coli paused disome complex (rotated-PRE-1 | closed non-rotated PRE)  EMDB-17288: Escherichia coli paused disome complex (rotated-PRE-1 | rotated PRE)  EMDB-17289: Cryo-EM reconstruction of the E. coli disome complex (rotated-PRE-1 | POST)  EMDB-17291: Escherichia coli paused disome complex (rotated-PRE-2 | closed non-rotated PRE)  EMDB-17431: Escherichia coli paused disome complex (rotated-PRE-2 | rotated PRE)  EMDB-17432: Escherichia coli paused disome complex (rotated-PRE-2 | POST)  EMDB-17433: Escherichia coli paused disome complex (POST | closed non-rotated PRE)  EMDB-17434: Escherichia coli paused disome complex (POST | rotated PRE)  EMDB-17441: Escherichia coli paused disome complex (POST | POST)  EMDB-17442: Escherichia coli paused disome complex (termination | closed non-rotated PRE)  EMDB-17443: Escherichia coli paused disome complex (termination | rotated PRE)  EMDB-17444: Escherichia coli paused disome complex (termination | POST) EMDB-17631, PDB-8peg: EMDB-17743, PDB-8pkl: EMDB-18875, PDB-8r3v: EMDB-19054, PDB-8rcl: EMDB-19055, PDB-8rcm: EMDB-19058, PDB-8rcs: EMDB-19059, PDB-8rct: 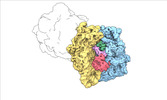 EMDB-19094: Escherichia coli paused disome complex (local refinement of leading decoding 70S) 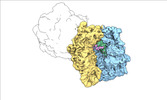 EMDB-19095: Escherichia coli paused disome complex (local refinement of leading 70S partially accommodated A-tRNA) 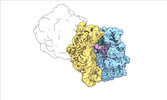 EMDB-19096: Escherichia coli paused disome complex (local refinement of leading 70S rotated PRE-1)  EMDB-19097: Escherichia coli paused disome complex (local refinement of leading 70S rotated PRE-2) 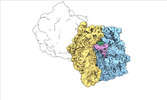 EMDB-19098: Escherichia coli paused disome complex (local refinement of leading 70S open non-rotated PRE) 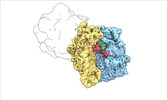 EMDB-19099: Escherichia coli paused disome complex (local refinement of leading 70S termination) 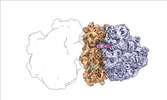 EMDB-19100: Escherichia coli paused disome complex (local refinement of queueing 70S rotated PRE-2) 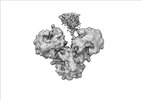 EMDB-19101: Escherichia coli paused trisome complex (non-rotated) 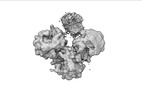 EMDB-19102: Escherichia coli paused trisome complex (rotated)  EMDB-19103: Escherichia coli paused disome complex (local refinement of queueing 70S POST)  EMDB-19104: Escherichia coli paused disome complex (local refinement of leading 70S POST) |
| Chemicals |  ChemComp-ZN:  ChemComp-MG:  ChemComp-ATP:  ChemComp-PUT:  ChemComp-SPD:  ChemComp-ALA:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords |  RIBOSOME / RIBOSOME /  translation / disome / translation / disome /  collision / PURE system / collision / PURE system /  polysome / tranlsation / elongation / pausing polysome / tranlsation / elongation / pausing |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers