+Search query
-Structure paper
| Title | Organization of an activator-bound RNA polymerase holoenzyme. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 32, Issue 3, Page 337-346, Year 2008 |
| Publish date | Nov 7, 2008 |
 Authors Authors | Daniel Bose / Tillmann Pape / Patricia C Burrows / Mathieu Rappas / Siva R Wigneshweraraj / Martin Buck / Xiaodong Zhang /  |
| PubMed Abstract | Transcription initiation involves the conversion from closed promoter complexes, comprising RNA polymerase (RNAP) and double-stranded promoter DNA, to open complexes, in which the enzyme is able to ...Transcription initiation involves the conversion from closed promoter complexes, comprising RNA polymerase (RNAP) and double-stranded promoter DNA, to open complexes, in which the enzyme is able to access the DNA template in a single-stranded form. The complex between bacterial RNAP and its major variant sigma factor sigma(54) remains as a closed complex until ATP hydrolysis-dependent remodeling by activator proteins occurs. This remodeling facilitates DNA melting and allows the transition to the open complex. Here we present cryoelectron microscopy reconstructions of bacterial RNAP in complex with sigma(54) alone, and of RNAP-sigma(54) with an AAA+ activator. Together with photo-crosslinking data that establish the location of promoter DNA within the complexes, we explain why the RNAP-sigma(54) closed complex is unable to access the DNA template and propose how the structural changes induced by activator binding can initiate conformational changes that ultimately result in formation of the open complex. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:18995832 / PubMed:18995832 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 21.0 - 23.0 Å |
| Structure data | 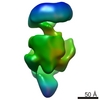 EMDB-1566: 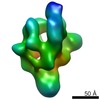 EMDB-1567:  EMDB-1568: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Escherichia coli (E. coli)
Escherichia coli (E. coli)