+Search query
-Structure paper
| Title | IFITM3 blocks influenza virus entry by sorting lipids and stabilizing hemifusion. |
|---|---|
| Journal, issue, pages | Cell Host Microbe, Vol. 31, Issue 4, Page 616-633.e20, Year 2023 |
| Publish date | Apr 12, 2023 |
 Authors Authors | Steffen Klein / Gonen Golani / Fabio Lolicato / Carmen Lahr / Daniel Beyer / Alexia Herrmann / Moritz Wachsmuth-Melm / Nina Reddmann / Romy Brecht / Mehdi Hosseinzadeh / Androniki Kolovou / Jana Makroczyova / Sarah Peterl / Martin Schorb / Yannick Schwab / Britta Brügger / Walter Nickel / Ulrich S Schwarz / Petr Chlanda /   |
| PubMed Abstract | Interferon-induced transmembrane protein 3 (IFITM3) inhibits the entry of numerous viruses through undefined molecular mechanisms. IFITM3 localizes in the endosomal-lysosomal system and specifically ...Interferon-induced transmembrane protein 3 (IFITM3) inhibits the entry of numerous viruses through undefined molecular mechanisms. IFITM3 localizes in the endosomal-lysosomal system and specifically affects virus fusion with target cell membranes. We found that IFITM3 induces local lipid sorting, resulting in an increased concentration of lipids disfavoring viral fusion at the hemifusion site. This increases the energy barrier for fusion pore formation and the hemifusion dwell time, promoting viral degradation in lysosomes. In situ cryo-electron tomography captured IFITM3-mediated arrest of influenza A virus membrane fusion. Observation of hemifusion diaphragms between viral particles and late endosomal membranes confirmed hemifusion stabilization as a molecular mechanism of IFITM3. The presence of the influenza fusion protein hemagglutinin in post-fusion conformation close to hemifusion sites further indicated that IFITM3 does not interfere with the viral fusion machinery. Collectively, these findings show that IFITM3 induces lipid sorting to stabilize hemifusion and prevent virus entry into target cells. |
 External links External links |  Cell Host Microbe / Cell Host Microbe /  PubMed:37003257 PubMed:37003257 |
| Methods | EM (tomography) |
| Structure data | 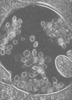 EMDB-15130: Tomogram of a late endosome of A549-IFITM3 cells infected with influenza A virus (Figure 4E)  EMDB-15131: Tomogram of a late endosome of A549-IFITM3 cells infected with influenza A virus (Figure S7) 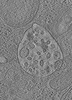 EMDB-15132: Tomogram of a late endosome of A549-IFITM3 cells infected with influenza A virus (Figure S8)  EMDB-15133: Tomogram of a late endosome of A549-IFITM3 cells infected with influenza A virus (Figure S9)  EMDB-15705: Tomogram of a late endosome of A549 cell (Figure 1R)  EMDB-15707: Tomogram of a late endosome of A549 cell treated with IFN-beta (Figure 1T) 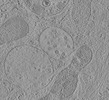 EMDB-15708: Tomogram of a late endosome of A549-IFITM3 cell (Figure 1V)  EMDB-16128: Tomogram of a late endosome of A549 cell infected with influenza A virus.  EMDB-16129: Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6C).  EMDB-16130: Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6E)  EMDB-16131: Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6G)  EMDB-16132: Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6I,K,M)  EMDB-16133: Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6O) |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Homo sapiens (human)
Homo sapiens (human)