+Search query
-Structure paper
| Title | Conformational coupling of redox-driven Na-translocation in Vibrio cholerae NADH:quinone oxidoreductase. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 30, Issue 11, Page 1686-1694, Year 2023 |
| Publish date | Sep 14, 2023 |
 Authors Authors | Jann-Louis Hau / Susann Kaltwasser / Valentin Muras / Marco S Casutt / Georg Vohl / Björn Claußen / Wojtek Steffen / Alexander Leitner / Eckhard Bill / George E Cutsail / Serena DeBeer / Janet Vonck / Julia Steuber / Günter Fritz /   |
| PubMed Abstract | In the respiratory chain, NADH oxidation is coupled to ion translocation across the membrane to build up an electrochemical gradient. In the human pathogen Vibrio cholerae, the sodium-pumping NADH: ...In the respiratory chain, NADH oxidation is coupled to ion translocation across the membrane to build up an electrochemical gradient. In the human pathogen Vibrio cholerae, the sodium-pumping NADH:quinone oxidoreductase (Na-NQR) generates a sodium gradient by a so far unknown mechanism. Here we show that ion pumping in Na-NQR is driven by large conformational changes coupling electron transfer to ion translocation. We have determined a series of cryo-EM and X-ray structures of the Na-NQR that represent snapshots of the catalytic cycle. The six subunits NqrA, B, C, D, E, and F of Na-NQR harbor a unique set of cofactors that shuttle the electrons from NADH twice across the membrane to quinone. The redox state of a unique intramembranous [2Fe-2S] cluster orchestrates the movements of subunit NqrC, which acts as an electron transfer switch. We propose that this switching movement controls the release of Na from a binding site localized in subunit NqrB. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:37710014 / PubMed:37710014 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.5 - 3.5 Å |
| Structure data | EMDB-15088, PDB-8a1t: EMDB-15089, PDB-8a1u: EMDB-15090, PDB-8a1v: EMDB-15091, PDB-8a1w: EMDB-15092, PDB-8a1x: EMDB-15093, PDB-8a1y:  PDB-8acw:  PDB-8acy:  PDB-8ad3:  PDB-8ad4:  PDB-8ad5: |
| Chemicals |  ChemComp-FMN:  ChemComp-RBF:  ChemComp-LMT:  ChemComp-3PE:  ChemComp-NA:  ChemComp-K:  ChemComp-FES:  ChemComp-FAD: 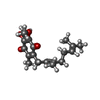 ChemComp-UQ2:  ChemComp-NAI:  ChemComp-HOH: 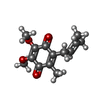 ChemComp-UQ1:  ChemComp-HQO:  ChemComp-LYS:  ChemComp-MG:  ChemComp-SO4: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  quinone / quinone /  NADH / sodium pump / NADH / sodium pump /  inhibitor / inhibitor /  respiratory complex / NADH ubiquinone oxido reducatase / Na+ pump / respiratory complex / NADH ubiquinone oxido reducatase / Na+ pump /  FLAVOPROTEIN / FAD / Na+-NQR FLAVOPROTEIN / FAD / Na+-NQR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers
















