+Search query
-Structure paper
| Title | Structural basis of GPBAR activation and bile acid recognition. |
|---|---|
| Journal, issue, pages | Nature, Vol. 587, Issue 7834, Page 499-504, Year 2020 |
| Publish date | Jul 22, 2020 |
 Authors Authors | Fan Yang / Chunyou Mao / Lulu Guo / Jingyu Lin / Qianqian Ming / Peng Xiao / Xiang Wu / Qingya Shen / Shimeng Guo / Dan-Dan Shen / Ruirui Lu / Linqi Zhang / Shenming Huang / Yuqi Ping / Chenlu Zhang / Cheng Ma / Kai Zhang / Xiaoying Liang / Yuemao Shen / Fajun Nan / Fan Yi / Vincent C Luca / Jiuyao Zhou / Changtao Jiang / Jin-Peng Sun / Xin Xie / Xiao Yu / Yan Zhang /   |
| PubMed Abstract | The G-protein-coupled bile acid receptor (GPBAR) conveys the cross-membrane signalling of a vast variety of bile acids and is a signalling hub in the liver-bile acid-microbiota-metabolism axis. Here ...The G-protein-coupled bile acid receptor (GPBAR) conveys the cross-membrane signalling of a vast variety of bile acids and is a signalling hub in the liver-bile acid-microbiota-metabolism axis. Here we report the cryo-electron microscopy structures of GPBAR-G complexes stabilized by either the high-affinity P395 or the semisynthesized bile acid derivative INT-777 at 3 Å resolution. These structures revealed a large oval pocket that contains several polar groups positioned to accommodate the amphipathic cholic core of bile acids, a fingerprint of key residues to recognize diverse bile acids in the orthosteric site, a putative second bile acid-binding site with allosteric properties and structural features that contribute to bias properties. Moreover, GPBAR undertakes an atypical mode of activation and G protein coupling that features a different set of key residues connecting the ligand-binding pocket to the G-coupling site, and a specific interaction motif that is localized in intracellular loop 3. Overall, our study not only reveals unique structural features of GPBAR that are involved in bile acid recognition and allosteric effects, but also suggests the presence of distinct connecting mechanisms between the ligand-binding pocket and the G-protein-binding site in the G-protein-coupled receptor superfamily. |
 External links External links |  Nature / Nature /  PubMed:32698187 PubMed:32698187 |
| Methods | EM (single particle) |
| Resolution | 3.0 Å |
| Structure data | EMDB-30344, PDB-7cfm: EMDB-30345, PDB-7cfn: |
| Chemicals | 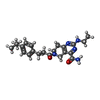 ChemComp-FWX:  ChemComp-CLR:  ChemComp-HOH: 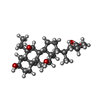 ChemComp-FX0:  ChemComp-PLM: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  GPCR / GPBAR / GPCR / GPBAR /  Complex / Complex /  Bile acid Bile acid |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








