+Search query
-Structure paper
| Title | Structural and functional insights into Escherichia coli α2-macroglobulin endopeptidase snap-trap inhibition. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 112, Issue 27, Page 8290-8295, Year 2015 |
| Publish date | Jul 7, 2015 |
 Authors Authors | Irene Garcia-Ferrer / Pedro Arêde / Josué Gómez-Blanco / Daniel Luque / Stephane Duquerroy / José R Castón / Theodoros Goulas / F Xavier Gomis-Rüth /   |
| PubMed Abstract | The survival of commensal bacteria requires them to evade host peptidases. Gram-negative bacteria from the human gut microbiome encode a relative of the human endopeptidase inhibitor, α2- ...The survival of commensal bacteria requires them to evade host peptidases. Gram-negative bacteria from the human gut microbiome encode a relative of the human endopeptidase inhibitor, α2-macroglobulin (α2M). Escherichia coli α2M (ECAM) is a ∼ 180-kDa multidomain membrane-anchored pan-peptidase inhibitor, which is cleaved by host endopeptidases in an accessible bait region. Structural studies by electron microscopy and crystallography reveal that this cleavage causes major structural rearrangement of more than half the 13-domain structure from a native to a compact induced form. It also exposes a reactive thioester bond, which covalently traps the peptidase. Subsequently, peptidase-laden ECAM is shed from the membrane and may dimerize. Trapped peptidases are still active except against very large substrates, so inhibition potentially prevents damage of large cell envelope components, but not host digestion. Mechanistically, these results document a novel monomeric "snap trap." |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:26100869 / PubMed:26100869 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.6 - 17.0 Å |
| Structure data | EMDB-3016, PDB-5a42: 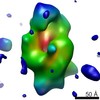 EMDB-3017:  EMDB-3018:  PDB-4ziq:  PDB-4ziu:  PDB-4zjg:  PDB-4zjh: |
| Chemicals |  ChemComp-GOL:  ChemComp-CL:  ChemComp-HOH:  ChemComp-NI:  ChemComp-PG4:  ChemComp-PGE:  ChemComp-1PE:  ChemComp-PEG:  ChemComp-ACT: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / Bacterial pan-proteinase inhibitor / MEMBRANE PROTEIN / Bacterial pan-proteinase inhibitor /  MEMBRANE PROTEIN/INHIBITOR / MEMBRANE PROTEIN/INHIBITOR /  membrane protein and inhibitor complex / membrane protein and inhibitor complex /  MEMBRANE PROTEIN-INHIBITOR complex / HYDROLASE INHIBITOR / MEMBRANE PROTEIN-INHIBITOR complex / HYDROLASE INHIBITOR /  PEPTIDASE INHIBITOR PEPTIDASE INHIBITOR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






