-Search query
-Search result
Showing 1 - 50 of 1,906 items for (Species: severe acute respiratory syndrome coronavirus 2) & (database: EMDB)

EMDB-36672: 
Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab

EMDB-40814: 
Local refinement of SARS-CoV-2 (HP-GSAS-Mut7) spike NTD in complex with TXG-0078 Fab

EMDB-16397: 
SARS-CoV2 Omicron BA.1 spike in complex with CAB-A17 antibody

EMDB-17296: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation

EMDB-39916: 
SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up)

EMDB-39917: 
SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up)

EMDB-39918: 
SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up)

EMDB-39919: 
SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region

EMDB-39921: 
SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up)

EMDB-39922: 
SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up)

EMDB-39923: 
SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region

EMDB-44644: 
SARS CoV2 spike in complex with NTD-directed antibody Fab DH1052
EMDB-18639: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)

EMDB-18649: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab

EMDB-19002: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
EMDB-36850: 
SARS-CoV-2 Omicron BA.1 spike protein in complex with a self-assembling trivalent nanobody Tr67
EMDB-34548: 
Spike 3-up RBD with THSC20.HVTR04 (Fab4): State - III
EMDB-34563: 
Spike 2-up RBD with THSC20.HVTR26 (Fab26): State - I

EMDB-34602: 
State - I: Spike 2-up RBD with THSC20.HVTR26 (Fab26)
EMDB-34603: 
State - II: Spike 3-up RBD with THSC20.HVTR26 (Fab26)
EMDB-34546: 
State - I: Spike 2-up RBD with THSC20.HVTR04 (Fab4)
EMDB-34547: 
Spike 3-up RBD with THSC20.HVTR04 (Fab4): State - II

EMDB-38453: 
Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state)

EMDB-38454: 
Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2

EMDB-38372: 
SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab

EMDB-42144: 
SARS-CoV-2 Nsp15, apo-form

EMDB-42145: 
SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form

EMDB-42146: 
SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1

EMDB-42147: 
SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2

EMDB-38284: 
XBB.1.5 spike protein in complex with BD55-1205

EMDB-37648: 
SARS-CoV-2 EG.5.1 spike glycoprotein (1-up state)

EMDB-37650: 
SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state)

EMDB-37651: 
SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state)
EMDB-37240: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab

EMDB-37241: 
The interface structure of Omicron RBD binding to 5817 Fab
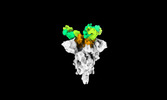
EMDB-35732: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with 1C4

EMDB-37626: 
Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with hippopotamus ACE2

EMDB-37629: 
Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with hippopotamus ACE2

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement

EMDB-40190: 
Local map of B3SB3L in complex with two-RBD-up state I of soluble SARS-CoV-2 Spike trimer
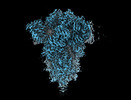
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
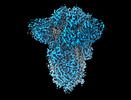
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein

EMDB-42820: 
SARS-CoV-2 5' proximal stem-loop 5 and 6 with SL5b extended

EMDB-29950: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10

EMDB-29975: 
Overall map of SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10

EMDB-40007: 
Local map of SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21B10

EMDB-18523: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes

EMDB-38283: 
XBB.1.5 RBD in complex with BD55-1205
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
