[English] 日本語
 Yorodumi
Yorodumi- EMDB-18639: Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2) | |||||||||
 Map data Map data | Sharpened refined map of BA-2.86 spike with ACE2. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Viral protein / Viral protein /  immune system / immune system /  SARS-CoV-2 / SARS-CoV-2 /  ACE2 / RBD / Spike / ACE2 / RBD / Spike /  glycoprotein / BA.2.86 / glycoprotein / BA.2.86 /  angiotensin converting enzyme 2 / angiotensin converting enzyme 2 /  receptor / receptor /  coronavirus-2 coronavirus-2 | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of amino acid transport /  angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane /  Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / tryptophan transport / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / tryptophan transport / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin /  regulation of vasoconstriction / regulation of cardiac conduction ...positive regulation of amino acid transport / regulation of vasoconstriction / regulation of cardiac conduction ...positive regulation of amino acid transport /  angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane /  Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / tryptophan transport / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / tryptophan transport / positive regulation of gap junction assembly / regulation of systemic arterial blood pressure by renin-angiotensin /  regulation of vasoconstriction / regulation of cardiac conduction / peptidyl-dipeptidase activity / angiotensin maturation / maternal process involved in female pregnancy / Metabolism of Angiotensinogen to Angiotensins / regulation of vasoconstriction / regulation of cardiac conduction / peptidyl-dipeptidase activity / angiotensin maturation / maternal process involved in female pregnancy / Metabolism of Angiotensinogen to Angiotensins /  metallocarboxypeptidase activity / Attachment and Entry / metallocarboxypeptidase activity / Attachment and Entry /  carboxypeptidase activity / negative regulation of signaling receptor activity / positive regulation of cardiac muscle contraction / regulation of cytokine production / carboxypeptidase activity / negative regulation of signaling receptor activity / positive regulation of cardiac muscle contraction / regulation of cytokine production /  viral life cycle / blood vessel diameter maintenance / brush border membrane / regulation of transmembrane transporter activity / negative regulation of smooth muscle cell proliferation / viral life cycle / blood vessel diameter maintenance / brush border membrane / regulation of transmembrane transporter activity / negative regulation of smooth muscle cell proliferation /  cilium / negative regulation of ERK1 and ERK2 cascade / endocytic vesicle membrane / cilium / negative regulation of ERK1 and ERK2 cascade / endocytic vesicle membrane /  metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / virus receptor activity / regulation of cell population proliferation / metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / virus receptor activity / regulation of cell population proliferation /  regulation of inflammatory response / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / regulation of inflammatory response / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release /  endopeptidase activity / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / endopeptidase activity / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / Potential therapeutics for SARS / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / symbiont entry into host cell / receptor ligand activity / host cell surface receptor binding / symbiont entry into host cell /  membrane raft / apical plasma membrane / fusion of virus membrane with host plasma membrane / membrane raft / apical plasma membrane / fusion of virus membrane with host plasma membrane /  endoplasmic reticulum lumen / fusion of virus membrane with host endosome membrane / endoplasmic reticulum lumen / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  cell surface / cell surface /  extracellular space / extracellular exosome / zinc ion binding / extracellular region / extracellular space / extracellular exosome / zinc ion binding / extracellular region /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Ren J / Stuart DI / Duyvesteyn HME | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2024 Journal: Cell Rep Med / Year: 2024Title: A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity. Authors: Chang Liu / Daming Zhou / Aiste Dijokaite-Guraliuc / Piyada Supasa / Helen M E Duyvesteyn / Helen M Ginn / Muneeswaran Selvaraj / Alexander J Mentzer / Raksha Das / Thushan I de Silva / ...Authors: Chang Liu / Daming Zhou / Aiste Dijokaite-Guraliuc / Piyada Supasa / Helen M E Duyvesteyn / Helen M Ginn / Muneeswaran Selvaraj / Alexander J Mentzer / Raksha Das / Thushan I de Silva / Thomas G Ritter / Megan Plowright / Thomas A H Newman / Lizzie Stafford / Barbara Kronsteiner / Nigel Temperton / Yuan Lui / Martin Fellermeyer / Philip Goulder / Paul Klenerman / Susanna J Dunachie / Michael I Barton / Mikhail A Kutuzov / Omer Dushek / / Elizabeth E Fry / Juthathip Mongkolsapaya / Jingshan Ren / David I Stuart / Gavin R Screaton /    Abstract: BA.2.86, a recently described sublineage of SARS-CoV-2 Omicron, contains many mutations in the spike gene. It appears to have originated from BA.2 and is distinct from the XBB variants responsible ...BA.2.86, a recently described sublineage of SARS-CoV-2 Omicron, contains many mutations in the spike gene. It appears to have originated from BA.2 and is distinct from the XBB variants responsible for many infections in 2023. The global spread and plethora of mutations in BA.2.86 has caused concern that it may possess greater immune-evasive potential, leading to a new wave of infection. Here, we examine the ability of BA.2.86 to evade the antibody response to infection using a panel of vaccinated or naturally infected sera and find that it shows marginally less immune evasion than XBB.1.5. We locate BA.2.86 in the antigenic landscape of recent variants and look at its ability to escape panels of potent monoclonal antibodies generated against contemporary SARS-CoV-2 infections. We demonstrate, and provide a structural explanation for, increased affinity of BA.2.86 to ACE2, which may increase transmissibility. #1:  Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2019 Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2019Title: Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix Authors: Liebschner D / Afonine PV #2:  Journal: Nature Methods / Year: 2017 Journal: Nature Methods / Year: 2017Title: cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination Authors: Punjani A / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18639.map.gz emd_18639.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18639-v30.xml emd-18639-v30.xml emd-18639.xml emd-18639.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18639.png emd_18639.png | 56.8 KB | ||
| Filedesc metadata |  emd-18639.cif.gz emd-18639.cif.gz | 7.4 KB | ||
| Others |  emd_18639_half_map_1.map.gz emd_18639_half_map_1.map.gz emd_18639_half_map_2.map.gz emd_18639_half_map_2.map.gz | 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18639 http://ftp.pdbj.org/pub/emdb/structures/EMD-18639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18639 | HTTPS FTP |
-Related structure data
| Related structure data |  8qsqMC  8qrfC  8qrgC  8qtdC  8r80C  8r8kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18639.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18639.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened refined map of BA-2.86 spike with ACE2. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A of BA-2.86 spike with ACE2.
| File | emd_18639_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of BA-2.86 spike with ACE2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
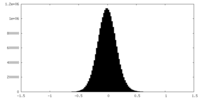
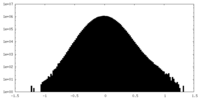
| Density Histograms |
-Half map: Half map B of BA-2.86 spike with ACE2.
| File | emd_18639_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of BA-2.86 spike with ACE2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
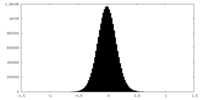
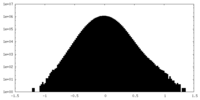
| Density Histograms |
- Sample components
Sample components
-Entire : BA-2.86 variant SARS-CoV2 S protein complexed with angiotensin co...
| Entire | Name: BA-2.86 variant SARS-CoV2 S protein complexed with angiotensin converting enzyme 2 (ACE2) |
|---|---|
| Components |
|
-Supramolecule #1: BA-2.86 variant SARS-CoV2 S protein complexed with angiotensin co...
| Supramolecule | Name: BA-2.86 variant SARS-CoV2 S protein complexed with angiotensin converting enzyme 2 (ACE2) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Spike protein recombinantly expressed using sequence of human-derived BA-2.86 variant of SARS-CoV2. Angiotensin converting enzyme 2 (ACE2) recombinantely expressed. |
|---|
-Supramolecule #2: Angiotensin converting enzyme 2 (ACE2)
| Supramolecule | Name: Angiotensin converting enzyme 2 (ACE2) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 Details: Human Angiotensin converting enzyme 2 (hACE2) recombinantely expressed |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: BA-2.86 variant SARS-CoV2 S protein
| Supramolecule | Name: BA-2.86 variant SARS-CoV2 S protein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 Details: Spike protein recombinantly expressed using sequence of human-derived BA-2.86 variant of SARS-CoV2. |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Macromolecule #1: Spike protein S2'
| Macromolecule | Name: Spike protein S2' / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Molecular weight | Theoretical: 22.159053 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VTNLCPFHEV FNATRFASVY AWNRTRISNC VADYSVLYNF APFFAFKCYG VSPTKLNDLC FTNVYADSFV IKGNEVSQIA PGQTGNIAD YNYKLPDDFT GCVIAWNSNK LDSKHSGNYD YWYRLFRKSK LKPFERDIST EIYQAGNKPC KGKGPNCYFP L QSYGFRPT ...String: VTNLCPFHEV FNATRFASVY AWNRTRISNC VADYSVLYNF APFFAFKCYG VSPTKLNDLC FTNVYADSFV IKGNEVSQIA PGQTGNIAD YNYKLPDDFT GCVIAWNSNK LDSKHSGNYD YWYRLFRKSK LKPFERDIST EIYQAGNKPC KGKGPNCYFP L QSYGFRPT YGVGHQPYRV VVLSFELLHA PATVCGP UniProtKB:  Spike glycoprotein Spike glycoprotein |
-Macromolecule #2: Processed angiotensin-converting enzyme 2
| Macromolecule | Name: Processed angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.982562 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STIEEQAKTF LDKFNHEAED LFYQSSLASW NYNTNITEEN VQNMNNAGDK WSAFLKEQST LAQMYPLQEI QNLTVKLQLQ ALQQNGSSV LSEDKSKRLN TILNTMSTIY STGKVCNPDN PQECLLLEPG LNEIMANSLD YNERLWAWES WRSEVGKQLR P LYEEYVVL ...String: STIEEQAKTF LDKFNHEAED LFYQSSLASW NYNTNITEEN VQNMNNAGDK WSAFLKEQST LAQMYPLQEI QNLTVKLQLQ ALQQNGSSV LSEDKSKRLN TILNTMSTIY STGKVCNPDN PQECLLLEPG LNEIMANSLD YNERLWAWES WRSEVGKQLR P LYEEYVVL KNEMARANHY EDYGDYWRGD YEVNGVDGYD YSRGQLIEDV EHTFEEIKPL YEHLHAYVRA KLMNAYPSYI SP IGCLPAH LLGDMWGRFW TNLYSLTVPF GQKPNIDVTD AMVDQAWDAQ RIFKEAEKFF VSVGLPNMTQ GFWENSMLTD PGN VQKAVC HPTAWDLGKG DFRILMCTKV TMDDFLTAHH EMGHIQYDMA YAAQPFLLRN GANEGFHEAV GEIMSLSAAT PKHL KSIGL LSPDFQEDNE TEINFLLKQA LTIVGTLPFT YMLEKWRWMV FKGEIPKDQW MKKWWEMKRE IVGVVEPVPH DETYC DPAS LFHVSNDYSF IRYYTRTLYQ FQFQEALCQA AKHEGPLHKC DISNSTEAGQ KLFNMLRLGK SEPWTLALEN VVGAKN MNV RPLLNYFEPL FTWLKDQNKN SFVGWSTDWS PYADHHHHHH UniProtKB:  Angiotensin-converting enzyme 2 Angiotensin-converting enzyme 2 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | BA-2.86 Spike with angiotensin converting enzyme 2 (ACE2) at a 6-fold excess of S protein. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 9818 / Average electron dose: 50.0 e/Å2 / Details: Images were collected in EER format. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 504319 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: Initial RBD-Spike from related deposition. ACE2 model. Also compared with 8IOV. |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final 3D classification | Number classes: 5 / Software - Name: cryoSPARC (ver. v.4.3.1) Details: Following classification into 8 classes, classification without alignment, focussing on RBD/ACE2 region. |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. v.4.3.1) |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. v.4.3.1) / Details: CryoSPARC. / Number images used: 15883 |
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Phenix real space refinement with manual checks using coot. | ||||||||
| Refinement | Space: REAL / Protocol: OTHER | ||||||||
| Output model |  PDB-8qsq: |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X