-Search query
-Search result
Showing all 42 items for (author: takeda & h)

EMDB-35912: 
Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex

EMDB-35926: 
Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA

EMDB-35965: 
Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA

PDB-8j12: 
Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex

PDB-8j1j: 
Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA

PDB-8j3r: 
Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA

EMDB-32019: 
Cryo-EM structure of SAM-Tom40 intermediate complex

PDB-7vku: 
Cryo-EM structure of SAM-Tom40 intermediate complex
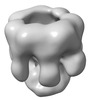
EMDB-32064: 
The oligomerized complex of hemolysin AF3 mutant protomers.
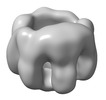
EMDB-32065: 
The oligomerized complex of AG2 hemolysin mutant protomers

EMDB-32118: 
Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex

PDB-7vtn: 
Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex

EMDB-30631: 
The mouse nucleosome structure containing H3mm18

EMDB-31882: 
The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv

PDB-7dbh: 
The mouse nucleosome structure containing H3mm18

PDB-7vbm: 
The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv

EMDB-30353: 
Split conformation 2 of CtHsp104 (Hsp104 from Chaetomium Thermophilum)

EMDB-30352: 
Split conformation 1 of CtHsp104 (Hsp104 from Chaetomium Thermophilum)

EMDB-30349: 
Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum)

PDB-7cg3: 
Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum)

EMDB-0869: 
Cryo-EM structure of the MgtE Mg2+ channel under Mg2+-free conditions

PDB-6lbh: 
Cryo-EM structure of the MgtE Mg2+ channel under Mg2+-free conditions

EMDB-30189: 
The mitochondrial SAM complex from S.cere

EMDB-30190: 
The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere

EMDB-30191: 
The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere

PDB-7btw: 
The mitochondrial SAM complex from S.cere

PDB-7btx: 
The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere

PDB-7bty: 
The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere

EMDB-30299: 
Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex

PDB-7c7l: 
Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex

EMDB-9618: 
Localization of FAP70 N-terminus on Chlamydomonas axoneme

EMDB-6844: 
Cofilin decorated actin filament

PDB-5yu8: 
Cofilin decorated actin filament

EMDB-6954: 
Doublet microtubule of zebrafish sperm axoneme, WT

EMDB-6955: 
Doublet microtubule of zebrafish sperm axoneme, pih1d1_null mutant

EMDB-6956: 
Doublet microtubule of zebrafish sperm axoneme, pih1d2_null mutant

EMDB-6957: 
Doublet microtubule of zebrafish sperm axoneme, ktu_null mutant

EMDB-6958: 
Doublet microtubule of zebrafish sperm axoneme, twister_null mutant

EMDB-6959: 
Doublet microtubule of zebrafish sperm axoneme, pih1d2_null and ktu_null double_mutant, +OAD

EMDB-6960: 
Doublet microtubule of zebrafish sperm axoneme, pih1d2_null and ktu_null double_mutant, -OAD

PDB-3iyo: 
Cryo-EM model of virion-sized HEV virion-sized capsid

EMDB-5173: 
Cryo-EM structure of virion-sized hepatitis E virus-like particle
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
