-Search query
-Search result
Showing 1 - 50 of 2,389 items for (author: mark & a)

EMDB-43592: 
PDI-containing spoke of a hexagonal wireframe DNA origami

EMDB-40261: 
DDB1/CRBN in complex with ARV-471 and the ER ligand-binding domain

EMDB-40815: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017

EMDB-40816: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 03

EMDB-40817: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 07

EMDB-40818: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp120-GH and base from participant 09

EMDB-40819: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, gp41-GH/FP and base from participant 11
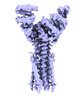
EMDB-50106: 
Artificial membrane protein TMHC4_R (ROCKET)
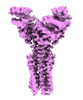
EMDB-50107: 
Artificial membrane protein TMHC4_R (ROCKET) mutant R9A/K10A/R13A

EMDB-19189: 
Human RAD52 closed ring

EMDB-19193: 
Human RAD52 open ring conformation

EMDB-19253: 
Human RAD52 open ring - ssDNA complex

EMDB-19255: 
Human Replication protein A (RPA; trimeric core) - ssDNA complex

PDB-8ril: 
Human RAD52 closed ring conformation

PDB-8rj3: 
Human RAD52 open ring conformation

PDB-8rjw: 
Human RAD52 open ring - ssDNA complex

PDB-8rk2: 
Human Replication protein A (RPA; trimeric core) - ssDNA complex

EMDB-17626: 
E. coli RNA polymerase paused at ops site

EMDB-17632: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site (alternative state of RfaH)

EMDB-17646: 
transcription complex paused at ops site and bound to autoinhibited RfaH, not fully complementary scaffold

EMDB-17647: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site (not fully complementary scaffold; alternative state of RfaH)

EMDB-17657: 
E. coli RNA polymerase paused at ops site (non-complementary scaffold)

EMDB-17668: 
E. coli transcription complex paused at ops site with fully recruited RfaH

EMDB-17679: 
autoinhibited RfaH bound to E. coli transcription complex paused at ops site (encounter complex)

EMDB-17681: 
backtracked E. coli transcription complex paused at ops site and bound to RfaH

EMDB-17685: 
E. coli transcription complex paused at ops site and bound to RfaH and NusA

EMDB-17686: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site (not complementary scaffold)

PDB-8pdy: 
E. coli RNA polymerase paused at ops site

PDB-8pen: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site (alternative state of RfaH)

PDB-8pfg: 
autoinhibited RfaH bound to E. coli transcription complex paused at ops site (encounter complex), not fully complementary scaffold

PDB-8pfj: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site (not fully complementary scaffold; alternative state of RfaH)

PDB-8ph9: 
E. coli RNA polymerase paused at ops site (non-complementary scaffold)

PDB-8phk: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site

PDB-8pib: 
autoinhibited RfaH bound to E. coli transcription complex paused at ops site (encounter complex)

PDB-8pid: 
backtracked E. coli transcription complex paused at ops site and bound to RfaH

PDB-8pil: 
E. coli transcription complex paused at ops site and bound to RfaH and NusA

PDB-8pim: 
fully recruited RfaH bound to E. coli transcription complex paused at ops site (not complementary scaffold)

EMDB-17870: 
Structure of the transcription termination factor Rho bound to RNA at the PBS and SBS

EMDB-17874: 
Structure of the transcription termination factor Rho in complex with Rof and ADP
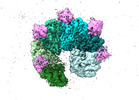
EMDB-17875: 
Structure of the transcription termination factor Rho in complex with Rof

EMDB-17876: 
Structure of Rho pentamer in complex with Rof and ADP
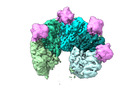
EMDB-17877: 
Structure of Rho pentamer in complex with Rof

PDB-8ptg: 
Structure of the transcription termination factor Rho bound to RNA at the PBS and SBS

PDB-8ptm: 
Structure of the transcription termination factor Rho in complex with Rof and ADP

PDB-8ptn: 
Structure of the transcription termination factor Rho in complex with Rof

PDB-8pto: 
Structure of Rho pentamer in complex with Rof and ADP

PDB-8ptp: 
Structure of Rho pentamer in complex with Rof
EMDB-41434: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
EMDB-41435: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
EMDB-41436: 
Central rod disk in D3 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
