-Search query
-Search result
Showing 1 - 50 of 67 items for (author: zhiqiang & chen)

EMDB-35155: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab
Method: single particle / : Li Z, Yu F, Cao S, ZHao H

EMDB-35156: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab
Method: single particle / : Li Z, Yu F, Cao S

PDB-8i3s: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab
Method: single particle / : Li Z, Yu F, Cao S, ZHao H

PDB-8i3u: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab
Method: single particle / : Li Z, Yu F, Cao S

EMDB-34084: 
Cryo-EM Structure of FGF23-FGFR4-aKlotho-HS Quaternary Complex
Method: single particle / : Mohammadi M, Chen L

PDB-7ysw: 
Cryo-EM Structure of FGF23-FGFR4-aKlotho-HS Quaternary Complex
Method: single particle / : Mohammadi M, Chen L

EMDB-35410: 
Arabinosyltransferase AftA
Method: single particle / : Gong YC, Rao ZH, Zhang L

EMDB-34075: 
Cryo-EM Structure of FGF23-FGFR1c-aKlotho-HS Quaternary Complex
Method: single particle / : Mohammadi M, Chen L

EMDB-34082: 
Cryo-EM Structure of FGF23-FGFR3c-aKlotho-HS Quaternary Complex
Method: single particle / : Mohammadi M, Chen L

PDB-7ysh: 
Cryo-EM Structure of FGF23-FGFR1c-aKlotho-HS Quaternary Complex
Method: single particle / : Mohammadi M, Chen L

PDB-7ysu: 
Cryo-EM Structure of FGF23-FGFR3c-aKlotho-HS Quaternary Complex
Method: single particle / : Mohammadi M, Chen L

EMDB-31623: 
Cryo EM structure of lysosomal ATPase
Method: single particle / : Zhang SS

EMDB-31626: 
Cryo EM structure of lysosomal ATPase
Method: single particle / : Zhang SS

EMDB-31627: 
Cryo EM structure of lysosomal ATPase
Method: single particle / : Zhang SS

EMDB-26200: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin
Method: single particle / : Kwon YD, Gorman J, Kwong PD
EMDB-26201: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin (Local refinement of FSR22 and RBD)
Method: single particle / : Kwon YD, Gorman J, Kwong PD
EMDB-27749: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022
Method: single particle / : Kwon YD, Gorman J, Kwong PD
EMDB-27750: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022
Method: single particle / : Kwon YD, Gorman J, Kwong PD

PDB-7tyz: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin
Method: single particle / : Kwon YD, Gorman J, Kwong PD

PDB-7tz0: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin (Local refinement of FSR22 and RBD)
Method: single particle / : Kwon YD, Gorman J, Kwong PD

PDB-8dw2: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022
Method: single particle / : Kwon YD, Gorman J, Kwong PD

PDB-8dw3: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022
Method: single particle / : Kwon YD, Gorman J, Kwong PD
EMDB-33019: 
SARS-CoV-2 BA.2 variant spike protein in complex with Fab BD55-5840
Method: single particle / : Wang X, Wang L
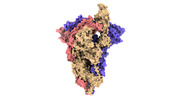
EMDB-33210: 
SARS-CoV-2 Omicron BA.2 variant spike (state 1)
Method: single particle / : Wang X, Wang L

EMDB-33211: 
SARS-CoV-2 Omicron BA.2 variant spike (state 2)
Method: single particle / : Wang X, Wang L
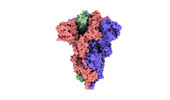
EMDB-33212: 
SARS-CoV-2 Omicron BA.3 variant spike
Method: single particle / : Wang X, Wang L
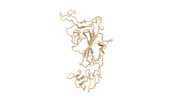
EMDB-33213: 
SARS-CoV-2 Omicron BA.3 variant spike (local)
Method: single particle / : Wang X, Wang L
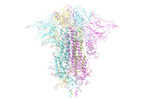
EMDB-33323: 
SARS-CoV-2 Omicron BA.4 variant spike
Method: single particle / : Wang X, Wang L
EMDB-33324: 
SARS-CoV-2 Omicron BA.2.13 variant spike
Method: single particle / : Wang X, Wang L
EMDB-33325: 
SARS-CoV-2 Omicron BA.2.12.1 variant spike
Method: single particle / : Wang X, Wang L

PDB-7x6a: 
SARS-CoV-2 BA.2 variant spike protein in complex with Fab BD55-5840
Method: single particle / : Wang X, Wang L

EMDB-32718: 
Local CryoEM structure of the SARS-CoV-2 S6P(B.1.1.529) in complex with BD55-3152 Fab
Method: single particle / : Du S, Xiao JY

EMDB-32732: 
Local structure of BD55-1239 Fab and SARS-COV2 Omicron RBD complex
Method: single particle / : Zhang ZZ, Xiao JJ

EMDB-32734: 
Local structure of BD55-3372 and delta spike
Method: single particle / : Liu PL

EMDB-32738: 
Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD
Method: single particle / : Zhang ZZ, Xiao JJ

PDB-7wr8: 
Local CryoEM structure of the SARS-CoV-2 S6P(B.1.1.529) in complex with BD55-3152 Fab
Method: single particle / : Du S, Xiao JY

PDB-7wrl: 
Local structure of BD55-1239 Fab and SARS-COV2 Omicron RBD complex
Method: single particle / : Zhang ZZ, Xiao JJ

PDB-7wrz: 
Local resolution of BD55-5840 Fab and SARS-COV2 Omicron RBD
Method: single particle / : Zhang ZZ, Xiao JJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search















 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
