-Search query
-Search result
Showing all 32 items for (author: yip & ck)

EMDB-37631: 
Hepatitis B virus capsid (HBV core protein)
Method: single particle / : Yip RPH, Lai LTF, Lau WCY, Ngo JCK, Kwok DCY

EMDB-37634: 
SR protein kinase 2 bound at 2-fold vertex of Hepatitis B virus capsid
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-38062: 
Hepatitis B virus capsid in complex with SR protein kinase 2
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-27738: 
Negative stain EM map of the heterodimeric p110gamma-p84 complex
Method: single particle / : Burke JE, Dalwadi U, Rathinaswamy MK, Yip CK, Nam SE

EMDB-27627: 
Structure of p110 gamma bound to the Ras inhibitory nanobody NB7
Method: single particle / : Burke JE, Nam SE, Rathinaswamy MK, Yip CK

EMDB-26792: 
Transcription co-activator SAGA
Method: single particle / : Vasyliuk D, Yip CK

EMDB-26803: 
Cryo-EM structure of the SAGA Tra1 module
Method: single particle / : Vasyliuk D, Yip CK

EMDB-26804: 
Cryo-EM structure of the SAGA core module
Method: single particle / : Vasyliuk D, Yip CK

EMDB-26616: 
SfSTING with c-di-GMP single fiber
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

EMDB-26617: 
SfSTING with c-di-GMP double fiber
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

EMDB-26618: 
SfSTING with cGAMP (masked)
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

EMDB-26619: 
SfSTING with cGAMP
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

PDB-7un8: 
SfSTING with c-di-GMP single fiber
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

PDB-7un9: 
SfSTING with c-di-GMP double fiber
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

PDB-7una: 
SfSTING with cGAMP (masked)
Method: single particle / : Morehouse BR, Yip MCJ, Keszei AFA, McNamara-Bordewick NK, Shao S, Kranzusch PJ

EMDB-23812: 
Structure of the apo phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex
Method: single particle / : Burke JE, Dalwadi U, Rathinaswamy MK, Yip CK

EMDB-23997: 
Structure of the mammalian Transport Protein Particle Complex (TRAPP) III
Method: single particle / : Harris NJ, Dalwadi U, Yip CK, Burke JE, Jenkins ML

EMDB-23808: 
Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex
Method: single particle / : Burke JE, Dalwadi U, Rathinaswamy MK, Yip CK

EMDB-23120: 
3D reconstruction of an autophagy tethering factor
Method: single particle / : Nam SE, Yip CK
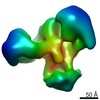
EMDB-7131: 
Chromatin-modifying complex
Method: single particle / : Setiaputra DT, Dalwadi U, Yip CK

EMDB-8239: 
EM map of the intact yeast Elongator complex
Method: single particle / : Setiaputra D, Cheng DTH, Hansen JM, Yip CK

EMDB-8291: 
EM map of the Elp123 subcomplex of yeast Elongator
Method: single particle / : Setiaputra D, Cheng DTH, Hansen JM, Yip CK

EMDB-6299: 
S. cerevisiae SAGA complex in the arched conformation
Method: single particle / : Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK

EMDB-6300: 
S. cerevisiae SAGA complex in the curved conformation
Method: single particle / : Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK

EMDB-6301: 
S. cerevisiae SAGA complex in the donut conformation
Method: single particle / : Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK

EMDB-6120: 
Structure of a mycobacterial protein
Method: single particle / : Solomonson M, Setiaputra D, Makepeace KAT, Lameignere E, Petrotchenko EV, Conrady DG, Bergeron JR, Vuckovic M, DiMaio F, Borchers CH, Yip CK, Strynadka NCJ

PDB-3j83: 
Heptameric EspB Rosetta model guided by EM density
Method: single particle / : Solomonson M, DiMaio F, Strynadka NCJ
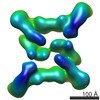
EMDB-5213: 
Molecular architecture of the yeast TRAPPII tethering complex - form1
Method: single particle / : Yip CK, Berscheminski J, Walz T

EMDB-5214: 
Molecular architecture of the yeast TRAPPII tethering complex - form2
Method: single particle / : Yip CK, Berscheminski J, Walz T
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
