-Search query
-Search result
Showing 1 - 50 of 59 items for (author: weimin & wu)

EMDB-26203: 
Structure of mitochondrial bc1 in complex with ck-2-68
Method: single particle / : Xia D, Esser L

PDB-7tz6: 
Structure of mitochondrial bc1 in complex with ck-2-68
Method: single particle / : Xia D, Esser L, Zhou F, Huang R

EMDB-29492: 
Three-Dimensional Reconstruction of HCV Envelope Glycoproteins E1E2 Heterodimer by Electron Microscopic Analysis
Method: single particle / : Kanai T, Liang TJ

EMDB-29499: 
Three-Dimensional Reconstruction of HCV Envelope Glycoproteins E1E2 Heterodimer by Electron Microscopic Analysis
Method: single particle / : Kanai T, Liang TJ

EMDB-25448: 
Negative-stain EM reconstruction of SpFN_1B-06-PL, a SARS-CoV-2 spike fused to H.pylori ferritin nanoparticle vaccine candidate
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25449: 
RFN_131, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Receptor-Binding Domain
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25450: 
pCoV146, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike Receptor-Binding and N-Terminal Domains
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25451: 
pCoV111, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike S1 Subunit
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-22331: 
SPN3US phage empty capsid
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22332: 
Phage SPN3US mottled capsid (glutaraldehyde-fixed)
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-22333: 
SPN3US phage mottled capsid
Method: single particle / : Heymann JB, Wang B, Newcomb WW, Wu W, Winkler DC, Cheng N, Reilly ER, Hsia RC, Thomas JA, Steven AC

EMDB-0849: 
cryo-EM structure of cyanobacteria Fd-NDH-1L complex
Method: single particle / : Zhang C, Shuai J

EMDB-0850: 
cryo-EM structure of cyanobacteria NDH-1LdelV complex
Method: single particle / : Zhang C, Shuai J

EMDB-0851: 
cryo-EM structure of cyanobacteria NDH-1LdelV complex
Method: single particle / : Zhang C, Shuai J, Wu J, Lei M

PDB-6l7o: 
cryo-EM structure of cyanobacteria Fd-NDH-1L complex
Method: single particle / : Zhang C, Shuai J, Wu J, Lei M

PDB-6l7p: 
cryo-EM structure of cyanobacteria NDH-1LdelV complex
Method: single particle / : Zhang C, Shuai J, Wu J, Lei M
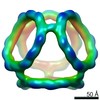
EMDB-21026: 
Truncated Tetrahedral RNA Nanostructures
Method: single particle / : Wu W, Zakrevsky P, Heinz W, Khant H, Kasprzak WK, Bindewald E, Dorjsuren N, Fields E, De Val N, Jaeger L, Shapiro BA

PDB-6ui6: 
HBV T=3 149C3A
Method: single particle / : Wu W, Watts NR, Cheng N, Huang R, Steven A, Wingfield PT

PDB-6ui7: 
HBV T=4 149C3A
Method: single particle / : Wu W, Watts NR, Cheng N, Huang R, Steven A, Wingfield PT

EMDB-20669: 
HBV T=3 particle
Method: single particle / : Wu W, Watts NR, Cheng N, Huang R, Steven A, Wingfield PT

EMDB-20670: 
HBV T=4 particle
Method: single particle / : Wu W

EMDB-8258: 
Reconstructions of P22 gp20 minus particles: low dose
Method: single particle / : Wu W, Leavitt JC

EMDB-8259: 
Reconstructions of P22 gp20 minus particles: 7th exposure
Method: single particle / : Wu W, Leavitt JC

EMDB-8260: 
Reconstructions of P22 gp20 minus particles: 8th exposure
Method: single particle / : Wu W, Leavitt JC

EMDB-8261: 
Reconstructions of P22 gp20 minus particles: 9th exposure
Method: single particle / : Wu W, Leavitt JC

EMDB-3358: 
HSV bubblegram imaging
Method: single particle / : Wu W, Newcomb WW, Cheng N, Aksyuk A, Winkler DC, Steven AC

EMDB-3359: 
HSV bubblegram imaging
Method: single particle / : Wu W, Newcomb WW, Cheng N, Aksyuk A, Winkler DC, Steven AC

EMDB-3360: 
HSV bubblegram imaging
Method: single particle / : Wu W, Newcomb WW, Cheng N, Aksyuk A, Winkler DC, Steven AC

EMDB-3361: 
HSV bubblegram imaging
Method: single particle / : Wu W, Newcomb WW, Cheng N, Aksyuk A, Winkler DC, Steven AC

EMDB-3288: 
HSV bubblegram imaging
Method: single particle / : Wu W, Newcomb WW, Cheng N, Aksyuk A, Winkler DC, Steven AC

EMDB-6034: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

EMDB-6035: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

EMDB-6036: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

EMDB-6037: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

PDB-3j7v: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

PDB-3j7w: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

PDB-3j7x: 
Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions
Method: single particle / : Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakata J, Chiu W, Serwer P, Jiang W

PDB-4btq: 
Coordinates of the bacteriophage phi6 capsid subunits fitted into the cryoEM map EMD-1206
Method: single particle / : Nemecek D, Boura E, Wu W, Cheng N, Plevka P, Qiao J, Mindich L, Heymann JB, Hurley JH, Steven AC

EMDB-2364: 
CryoEM reconstruction of the bacteriophage phi6 procapsid to the near-atomic resolution
Method: single particle / : Nemecek D, Boura E, Wu W, Cheng N, Plevka P, Qiao J, Mindich L, Heymann JB, Hurley JH, Steven AC

PDB-4btg: 
Coordinates of the bacteriophage phi6 capsid subunits (P1A and P1B) fitted into the cryoEM reconstruction of the procapsid at 4.4 A resolution
Method: single particle / : Nemecek D, Boura E, Wu W, Cheng N, Plevka P, Qiao J, Mindich L, Heymann JB, Hurley JH, Steven AC

EMDB-5678: 
Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling
Method: single particle / : Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W

PDB-3j40: 
Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling
Method: single particle / : Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W

EMDB-5566: 
Icosahedral reconstruction (map 1/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA Virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5567: 
Standard asymmetric reconstruction (map 2/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5568: 
Focused asymmetric reconstruction I (map 3/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5569: 
Focused asymmetric reconstruction II (map 4/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5570: 
Focused asymmetric reconstruction III (map 5/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5571: 
Focused asymmetric reconstruction IV (map 6/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5572: 
Focused asymmetric reconstruction V (map 7/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W

EMDB-5573: 
Focused asymmetric reconstruction VI (map 8/8): Visualization of Uncorrelated Tandem Symmetry Mismatches in the Internal Genome Packaging Apparatus of a dsDNA virus
Method: single particle / : Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright E, Serwer P, Jiang W
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
