-Search query
-Search result
Showing all 48 items for (author: toropova & k)

EMDB-19132: 
Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Mladenov M, Seda M, Jenkins D, Stephens DJ, Roberts AJ

EMDB-19133: 
Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Seda M, Jenkins D, Stephens DJ, Roberts AJ

PDB-8rgg: 
Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Mladenov M, Seda M, Jenkins D, Stephens DJ, Roberts AJ

PDB-8rgh: 
Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Seda M, Jenkins D, Stephens DJ, Roberts AJ

EMDB-15954: 
Structure of the IFT-A complex; IFT-A2 module
Method: single particle / : Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ

PDB-8bbe: 
Structure of the IFT-A complex; IFT-A2 module
Method: single particle / : Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ

PDB-8bbg: 
Structure of the IFT-A complex; anterograde IFT-A train model
Method: single particle / : Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ

EMDB-15955: 
Structure of the IFT-A complex; IFT-A1 module
Method: single particle / : Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ

PDB-8bbf: 
Structure of the IFT-A complex; IFT-A1 module
Method: single particle / : Hesketh SJ, Mukhopadhyay AG, Nakamura D, Toropova K, Roberts AJ

PDB-6sc2: 
Structure of the dynein-2 complex; IFT-train bound model
Method: single particle / : Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ

EMDB-4917: 
Structure of the dynein-2 complex; motor domains
Method: single particle / : Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ

EMDB-4918: 
Structure of the dynein-2 complex; tail domain
Method: single particle / : Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ

PDB-6rla: 
Structure of the dynein-2 complex; motor domains
Method: single particle / : Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ

PDB-6rlb: 
Structure of the dynein-2 complex; tail domain
Method: single particle / : Toropova K, Zalyte R, Mukhopadhyay AG, Mladenov M, Carter AP, Roberts AJ

EMDB-8484: 
Bacteriophage G
Method: single particle / : Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF

EMDB-8485: 
Bacteriophage 121Q
Method: single particle / : Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF

EMDB-8486: 
Bacillus phage PBS1
Method: single particle / : Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF

EMDB-8487: 
Bacteriophage N3
Method: single particle / : Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF

EMDB-8488: 
Bacteriophage PAU
Method: single particle / : Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF

EMDB-8489: 
Bacteriophage Bellamy
Method: single particle / : Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF

EMDB-6008: 
Dynein motor domain in complex with Lis1 in the absence of nucleotide
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE
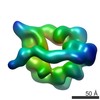
EMDB-6013: 
Dynein motor domain in the absence of nucleotide
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE

EMDB-6014: 
Dynein motor domain in the presence of ADP, with linker at position 2
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE
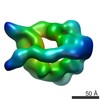
EMDB-6015: 
Dynein motor domain in the presence of ADP, with linker at position 1
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE
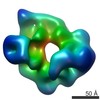
EMDB-6016: 
Dynein motor domain in complex with Lis1 in the presence of ATP and Vi
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE

EMDB-6017: 
Dynein motor domain with truncated linker in complex with Lis1 in the absence of nucleotide
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE

EMDB-5650: 
Pseudorabies virus C-capsid from cryo-electron microscopy
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5652: 
Pseudorabies virus B-capsid from cryo-electron microscopy
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5654: 
Pseudorabies virus A-capsid from cryo-electron microscopy
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5655: 
Pseudorabies virus virion capsid from cryo-electron microscopy
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5656: 
Pseudorabies GS-2168 B-capsid from cryo-electron microscopy
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5657: 
Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5659: 
Cryo-electron microscopy of herpesvirus simplex type 1 C-capsid
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5660: 
Cryo-electron microscopy of herpesvirus simplex type 1 B-capsid
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-5661: 
Cryo-electron microscopy of herpesvirus simplex type 1 A-capsid
Method: single particle / : Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF

EMDB-1902: 
The HSV-1 UL17 protein is the second constituent of the capsid vertex specific component (CVSC) required for DNA packaging and retention.
Method: single particle / : Toropova K, Huffman JB, Homa FL, Conway JF

EMDB-1903: 
The HSV-1 UL17 protein is the second constituent of the capsid vertex specific component (CVSC) required for DNA packaging and retention.
Method: single particle / : Toropova K, Huffman JB, Homa FL, Conway JF

EMDB-1905: 
Residues of the UL25 Protein of Herpes Simplex Virus That Are Required for Its Stable Interaction with Capsids.
Method: single particle / : Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL

EMDB-1879: 
Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid.
Method: single particle / : Rolfsson O, Toropova K, Ranson NA, Stockley PG

EMDB-1880: 
Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid.
Method: single particle / : Rolfsson O, Toropova K, Ranson NA, Stockley PG

EMDB-1881: 
Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid.
Method: single particle / : Rolfsson O, Toropova K, Ranson NA, Stockley PG

EMDB-1882: 
Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid.
Method: single particle / : Rolfsson O, Toropova K, Ranson NA, Stockley PG
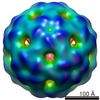
EMDB-1860: 
Visualising an RNA genome poised for release from its receptor complex.
Method: single particle / : Toropova K, Stockley PG, Ranson NA

EMDB-1859: 
Visualising a viral RNA genome poised for release from its receptor complex.
Method: single particle / : Toropova K, Stockley PG, Ranson NA

EMDB-1861: 
Visualising an RNA genome poised for release from its receptor complex.
Method: single particle / : Toropova K, Stockley PG, Ranson NA

EMDB-1431: 
The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly.
Method: single particle / : Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA

EMDB-1432: 
The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly.
Method: single particle / : Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA

EMDB-1433: 
The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly.
Method: single particle / : Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
