-Search query
-Search result
Showing all 17 items for (author: pandurangan & ap)

EMDB-15022: 
CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF
Method: single particle / : Hardwick SW, Kefala-Stavridi A, Chirgadze DY, Blundell TL, Chaplin AK

EMDB-16044: 
DNA-PK Ku80 mediated dimer bound to PAXX
Method: single particle / : Hardwick SW, Chaplin AK

EMDB-16070: 
DNA-PK XLF mediated dimer bound to PAXX
Method: single particle / : Hardwick SW, Chaplin AK

EMDB-16074: 
DNA-PK Ku80 mediated dimer bound to PAXX and XLF
Method: single particle / : Hardwick SW, Chaplin AK

PDB-7zyg: 
CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF
Method: single particle / : Hardwick SW, Kefala-Stavridi A, Chirgadze DY, Blundell TL, Chaplin AK

PDB-8bh3: 
DNA-PK Ku80 mediated dimer bound to PAXX
Method: single particle / : Hardwick SW, Chaplin AK

PDB-8bhv: 
DNA-PK XLF mediated dimer bound to PAXX
Method: single particle / : Hardwick SW, Chaplin AK

PDB-8bhy: 
DNA-PK Ku80 mediated dimer bound to PAXX and XLF
Method: single particle / : Hardwick SW, Chaplin AK

EMDB-14995: 
CryoEM structure of Ku heterodimer bound to DNA and PAXX
Method: single particle / : Hardwick SW, Kefala-Stavridi A, Chirgadze DY, Blundell TL, Chaplin AK

PDB-7zwa: 
CryoEM structure of Ku heterodimer bound to DNA and PAXX
Method: single particle / : Hardwick SW, Kefala-Stavridi A, Chirgadze DY, Blundell TL, Chaplin AK

EMDB-3362: 
Natively membrane-anchored full-length Herpes simplex virus 1 glycoprotein B
Method: subtomogram averaging / : Zeev-Ben-Mordehai T, Vasishtan D, Duran AH, Vollmer B, White P, Pandurangan AP, Siebert CA, Topf M, Grunewald K

EMDB-2979: 
Cryo EM structure of suilysin prepore
Method: single particle / : Dudkina NV, Leung C, Lukoyanova N, Hodel AW, Farabella I, Pandurangan AP, Jahan N, Damaso MP, Osmanovic D, Reboul CF, Dunstone MA, Andrew PW, Lonnen R, Topf M, Saibil HR, Hoogenboom BW

EMDB-2983: 
Cryo EM structure of suilysin pore
Method: single particle / : Dudkina NV, Leung C, Lukoyanova N, Hodel AW, Farabella I, Pandurangan AP, Jahan N, Pires Damaso M, Osmanovic D, Reboul CF, Dunstone MA, Andrew PW, Lonnen R, Topf M, Saibil HR, Hoogenboom BW
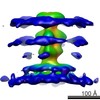
EMDB-2379: 
Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction
Method: subtomogram averaging / : Maurer UE, Zeev-Ben-Mordehai Z, Pandurangan AP, Cairns TM, Hannah BP, Whitbeck JC, Eisenberg RJ, Cohen GH, Topf M, Huiskonen JT, Grunewald K

EMDB-2380: 
Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction
Method: subtomogram averaging / : Maurer UE, Zeev-Ben-Mordehai Z, Pandurangan AP, Cairns TM, Hannah BP, Whitbeck JC, Eisenberg RJ, Cohen GH, Topf M, Huiskonen JT, Grunewald K

EMDB-2027: 
Coxsackievirus A7 (CAV7) empty capsid reconstruction at 6.09 angstrom resolution.
Method: single particle / : Seitsonen JJT, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ

EMDB-2028: 
Coxsackievirus A7 (CAV7) full capsid reconstruction at 8.23 angstrom resolution.
Method: single particle / : Seitsonen JJT, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
