-Search query
-Search result
Showing 1 - 50 of 399 items for (author: lok & s)

EMDB-36429: 
Cryo-EM structure of dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36430: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 4 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36431: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36432: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 4 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36433: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 37 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36434: 
Cryo-EM structure of the small tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C
Method: helical / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36435: 
Cryo-EM structure of the big tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C
Method: helical / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36436: 
Cryo-EM structure of dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36437: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 4 deg C
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36438: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36439: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 4 deg C
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36440: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 37 deg C
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-36441: 
Cryo-EM structure of dengue virus serotype 3 strain CH53489 spherical particle in complex with human antibody DENV-290 Fab at 37 deg C
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn1: 
Cryo-EM structure of dengue virus serotype 3 strain EHIE46200Y19 in complex with human antibody DENV-115 IgG at 4 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn2: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 4 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn3: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn4: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 4 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn5: 
Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 37 deg C (subparticle LLR-LRR)
Method: single particle / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn6: 
Cryo-EM structure of the small tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C
Method: helical / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

PDB-8jn7: 
Cryo-EM structure of the big tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C
Method: helical / : Fibriansah G, Ng TS, Tan AWK, Shi J, Lok SM

EMDB-19003: 
The structure of inactivated mature tick-borne encephalitis virus
Method: single particle / : Pichkur EB, Samygina VR

PDB-8r8l: 
The structure of inactivated mature tick-borne encephalitis virus
Method: single particle / : Pichkur EB, Samygina VR

EMDB-43629: 
Cryo-EM structure of phage DEV ejection proteins gp72:gp73
Method: single particle / : Iglesias SM, Cingolani G

PDB-8vxq: 
Cryo-EM structure of phage DEV ejection proteins gp72:gp73
Method: single particle / : Iglesias SM, Cingolani G

EMDB-16334: 
Cryo-EM structure of a Staphylococus aureus 30S-RbfA complex
Method: single particle / : Bikmullin AG, Fatkhullin B, Stetsenko A, Guskov A, Yusupov M
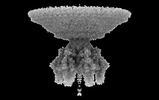
EMDB-41649: 
P22 Mature Virion tail - C6 Localized Reconstruction
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

EMDB-41651: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

EMDB-41819: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

PDB-8tvr: 
In situ cryo-EM structure of bacteriophage P22 tail hub protein: tailspike protein complex at 2.8A resolution
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

PDB-8tvu: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

PDB-8u1o: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

EMDB-41791: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

EMDB-41792: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

PDB-8u10: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

PDB-8u11: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

EMDB-34715: 
Cryo-EM structure of ComA bound to its mature substrate CSP peptide
Method: single particle / : Yu L, Xin X, Min L
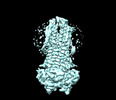
EMDB-36882: 
Cryo-EM structure of nucleotide-bound ComA with ZinC ion
Method: single particle / : Yu L, Xin X, Min L, Feng H

EMDB-36936: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant with Mg2+
Method: single particle / : Yu L, Xin X, Min L

PDB-8hf7: 
Cryo-EM structure of ComA bound to its mature substrate CSP peptide
Method: single particle / : Yu L, Xin X, Min L

PDB-8k4b: 
Cryo-EM structure of nucleotide-bound ComA with ZinC ion
Method: single particle / : Yu L, Xin X, Min L, Feng H

PDB-8k7a: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant with Mg2+
Method: single particle / : Yu L, Xin X, Min L

EMDB-16672: 
Pyruvate dehydrogenase complex core (E2, dihydrolipoyl transacetylase)
Method: subtomogram averaging / : Plokhikh KS, Chesnokov YM

EMDB-16731: 
Alpha-ketoglutarate dehydrogenase complex core (E2, dihydrolipoyl transsuccinylase), Branched-chain alpha-ketoacid dehydrogenase complex core (E2, dihydrolipoyl transacylase), indistinguishable
Method: subtomogram averaging / : Plokhikh KS, Chesnokov YM
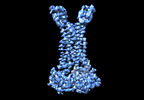
EMDB-35362: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-35363: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M
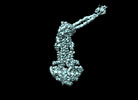
EMDB-35364: 
Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-35437: 
Cryo-EM structure of Mycobacterium tuberculosis ADP bound FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-36304: 
Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8idb: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8idc: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
