-Search query
-Search result
Showing all 30 items for (author: lauren & camp)

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
Method: single particle / : Duhoo Y, Lau K
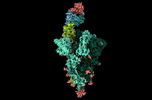
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)
Method: single particle / : Duhoo Y, Lau K

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.
Method: single particle / : Duhoo Y, Lau K

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-14141: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

EMDB-14142: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D
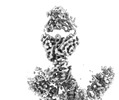
EMDB-14143: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qti: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qtj: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qtk: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

EMDB-13190: 
P5C3 is a potent fab neutralizer
Method: single particle / : perez L

EMDB-13265: 
the local resolution of Fab p5c3.
Method: single particle / : perez L

EMDB-13415: 
MaP OF P5C3RBD Interface
Method: single particle / : Perez L

EMDB-20024: 
Cryo-EM structure of Helicobacter pylori VacA hexamer
Method: single particle / : Erwin AL, Cover TL, Ohi MD

PDB-6ody: 
Cryo-EM structure of Helicobacter pylori VacA hexamer
Method: single particle / : Erwin AL, Cover TL, Ohi MD

EMDB-20029: 
Cryo-EM structure of Helicobacter pylori VacA heptamer
Method: single particle / : Erwin AL, Cover TL, Ohi MD

EMDB-5041: 
Ribosome structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5042: 
Lumazine synthase structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5043: 
GroEL structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5044: 
RNA polymerase structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5045: 
Phosphoenolpyruvate synthase structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5046: 
Putative protein structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5047: 
Inosine-5-monophosphate dehydrogenase structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
