-Search query
-Search result
Showing 1 - 50 of 224 items for (author: kuhn & r)

EMDB-13477: 
Small subunit of the Chlamydomonas reinhardtii mitoribosome
Method: single particle / : Waltz F, Soufari H, Hashem Y
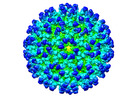
EMDB-26945: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ
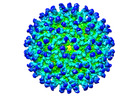
EMDB-26946: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ
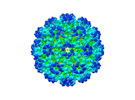
EMDB-26947: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

PDB-7v0n: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

PDB-7v0o: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

PDB-7v0p: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

EMDB-26794: 
Tomogram of platelet filopodia in presence of SARS-CoV-2 spike protein
Method: electron tomography / : Kuhn CC, Basnet N, Bodakuntla S, Nichols S, Martinez-Sanchez A, Agostini L, Soh YM, Takagi J, Biertumpfel C, Mizuno N

EMDB-26796: 
Tomogram of platelet protrusion with microtubule in presence of SARS-CoV-2 spike protein
Method: electron tomography / : Kuhn CC, Basnet N, Bodakuntla S, Nichols S, Martinez-Sanchez A, Agostini L, Soh YM, Takagi J, Biertumpfel C, Mizuno N

EMDB-26798: 
SARS-CoV-2 spike protein closed conformation
Method: single particle / : Kuhn CC, Basnet N, Bodakuntla S, Alvarez-Brecht P, Nichols S, Martinez-Sanchez A, Agostini L, Soh YM, Takagi J, Biertumpfel C, Mizuno N

EMDB-29020: 
Structure of dengue virus (DENV2) in complex with prM12, an anti-PrM monoclonal antibody
Method: single particle / : Dowd AD, Sirohi D, Speer S, Mukherjee S, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Kielian M, Kuhn RJ, Diamond MS, Pierson TC

EMDB-29021: 
Structure of dengue virus (DENV2) in complex with prM13, an anti-PrM monoclonal antibody
Method: single particle / : Dowd AD, Sirohi D, Speer S, Mukherjee S, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Kielian M, Kuhn RJ, Diamond MS, Pierson TC

PDB-8fe3: 
Structure of dengue virus (DENV2) in complex with prM12, an anti-PrM monoclonal antibody
Method: single particle / : Dowd AD, Sirohi D, Speer S, Mukherjee S, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Kielian M, Kuhn RJ, Diamond MS, Pierson TC

PDB-8fe4: 
Structure of dengue virus (DENV2) in complex with prM13, an anti-PrM monoclonal antibody
Method: single particle / : Dowd AD, Sirohi D, Speer S, Mukherjee S, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Kielian M, Kuhn RJ, Diamond MS, Pierson TC

EMDB-25765: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain native virion
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-25772: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with inhibitor 11526092
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-25773: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-25774: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain in complex with inhibitor 11526091 (no/low occupancy-no inhibitor modeled)
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-25776: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain in complex with inhibitor 11526093 (no/low occupancy-no inhibitor modeled)
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7t9p: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain native virion
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7taf: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with inhibitor 11526092
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7tag: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7tah: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain in complex with inhibitor 11526091 (no/low occupancy-no inhibitor modeled)
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7taj: 
Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain in complex with inhibitor 11526093 (no/low occupancy-no inhibitor modeled)
Method: single particle / : Fu J, Klose T, Kuhn RJ, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-14771: 
Amyloid fibril from human systemic AA amyloidosis (vascular variant)
Method: helical / : Banerjee S, Schmidt M, Faendrich M

PDB-7zky: 
Amyloid fibril from human systemic AA amyloidosis (vascular variant)
Method: helical / : Banerjee S, Schmidt M, Faendrich M
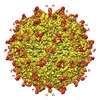
EMDB-25606: 
Zika Virus particle bound with IgM antibody DH1017 Fab fragment
Method: single particle / : Miller AS, Kuhn RJ

PDB-7t17: 
Zika Virus asymmetric unit bound with IgM antibody DH1017 Fab fragment
Method: single particle / : Miller AS, Kuhn RJ

EMDB-24606: 
EV-71
Method: single particle / : Chen CL, Kuhn RJ, Klose T

EMDB-24199: 
Localized reconstruction of EEEV
Method: single particle / : Chen CL, Kuhn RJ, Klose T

EMDB-24200: 
Pre-fusion state 2 of EEEV
Method: single particle / : Chen CL, Kuhn RJ, Klose T
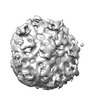
EMDB-24201: 
Pre-fusion state 2 of EEEV with localized reconstruction
Method: single particle / : Chen CL, Kuhn RJ, Klose T

EMDB-24202: 
EEEV in complex with Fab22 at acidic pH (pH 5.5)
Method: single particle / : Chen CL, Kuhn RJ, Klose T

EMDB-24203: 
EEEV in complex with Fab22 at neutral pH (pH 7.4)
Method: single particle / : Chen CL, Kuhn RJ, Klose T

EMDB-24204: 
Pre-fusion state 1 of EEEV
Method: single particle / : Chen CL, Kuhn RJ, Klose T
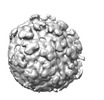
EMDB-24205: 
Pre-fusion state 1 of EEEV with localized reconstruction
Method: single particle / : Chen CL, Kuhn RJ, Klose T

PDB-7n69: 
Pre-fusion state 2 of EEEV with localized reconstruction
Method: single particle / : Chen CL, Kuhn RJ, Klose T
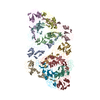
PDB-7n6a: 
Pre-fusion state 1 of EEEV with localized reconstruction
Method: single particle / : Chen CL, Kuhn RJ, Klose T

EMDB-13480: 
Large subunit of the Chlamydomonas reinhardtii mitoribosome
Method: single particle / : Waltz F, Soufari H, Hashem Y

PDB-7pkt: 
Large subunit of the Chlamydomonas reinhardtii mitoribosome
Method: single particle / : Waltz F, Soufari H, Hashem Y

EMDB-25471: 
Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound
Method: single particle / : Milligan JC, Yu X, Saphire EO

PDB-7swd: 
Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound
Method: single particle / : Milligan JC, Yu X, Saphire EO

EMDB-14185: 
Human RBBP6-bound CPSF complex
Method: single particle / : Sandmeir F, Conti E

EMDB-13481: 
Small subunit of the Chlamydomonas mitoribosome - head focus
Method: single particle / : Waltz F, Salinas-Giege T, Englmeier R, Meichel H, Soufari H, Kuhn L, Pfeffer S, Foerster F, Engel BD, Giege P, Drouard L, Hashem Y

EMDB-13578: 
C.reinhardtii mitochondrial ribosome (in situ)
Method: subtomogram averaging / : Waltz F, Salinas-Giege T, Englmeier R, Meichel H, Soufari H, Kuhn L, Pfeffer S, Foerster F, Engel BD, Giege P, Drouard L, Hashem Y

EMDB-11896: 
43S preinitiation complex from Leishmania tarentolae with kDDX60 helicase
Method: single particle / : Bochler A, Brito Querido J, Prilepskaja T, Soufari H, Del Cistia ML, Kuhn L, Rimoldi Ribeiro A, Valasek LS, Hashem Y

EMDB-13453: 
Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution.
Method: single particle / : Heyder NA, Schmidt A, Kleinau G, Hilal T, Scheerer P

EMDB-13454: 
Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution.
Method: single particle / : Heyder NA, Schmidt A, Kleinau G, Hilal T, Scheerer P

EMDB-22931: 
The capsid of Myoviridae Phage XM1
Method: single particle / : Wang Z, Klose T, Jiang W, Kuhn RJ

EMDB-22959: 
The Tail of Phage XM1
Method: single particle / : Wang Z, Klose T
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
