-Search query
-Search result
Showing 1 - 50 of 477 items for (author: jie & fu)

EMDB-41770: 
Apo form of human ATE1
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-42071: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

PDB-8uau: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-37210: 
Prefusion RSV F Bound to Lonafarnib and D25 Fab
Method: single particle / : Yang Q, Xue B, Liu F, Peng W, Chen X

PDB-8kg5: 
Prefusion RSV F Bound to Lonafarnib and D25 Fab
Method: single particle / : Yang Q, Xue B, Liu F, Peng W, Chen X

EMDB-37754: 
Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37755: 
Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37757: 
Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37758: 
Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-37759: 
Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqu: 
Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqv: 
Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqx: 
Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wqy: 
Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

PDB-8wr0: 
Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin
Method: single particle / : Wang WM, Ma DY, Gong WJ, Wu LJ, Wang HF

EMDB-38203: 
Cryo-EM structure of HerA
Method: single particle / : Wang Y, Deng Z

EMDB-38204: 
Cryo-EM structure of an anti-phage defense complex
Method: single particle / : Wang Y, Deng Z

EMDB-38205: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 1
Method: single particle / : An Q, Deng Z

EMDB-38206: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 2
Method: single particle / : An Q, Deng Z

EMDB-38207: 
Cryo-EM structure of an anti-phage defense complex bound to ATPrS and DNA
Method: single particle / : An Q, Deng Z

PDB-8xav: 
Cryo-EM structure of an anti-phage defense complex
Method: single particle / : Wang Y, Deng Z

PDB-8xaw: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 1
Method: single particle / : An Q, Deng Z

PDB-8xax: 
Cryo-EM structure of an anti-phage defense complex bound to AMPPNP and DNA at state 2
Method: single particle / : An Q, Deng Z

PDB-8xay: 
Cryo-EM structure of an anti-phage defense complex bound to ATPrS and DNA
Method: single particle / : An Q, Deng Z

EMDB-35827: 
Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37652: 
Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37656: 
Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary)
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37657: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary)
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-37762: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex)
Method: single particle / : Zhang S, Lin S, Liu JJG

PDB-8iyq: 
Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate
Method: single particle / : Zhang S, Lin S, Liu JJG

PDB-8wmh: 
Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate
Method: single particle / : Zhang S, Lin S, Liu JJG

PDB-8wmm: 
Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary)
Method: single particle / : Zhang S, Lin S, Liu JJG

PDB-8wmn: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary)
Method: single particle / : Zhang S, Lin S, Liu JJG

PDB-8wr4: 
Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex)
Method: single particle / : Zhang S, Lin S, Liu JJG

EMDB-38679: 
Cryo-EM structure of tomato NRC2 dimer
Method: single particle / : Sun Y, Ma SC, Chai JJ

EMDB-38680: 
Cryo-EM structure of tomato NRC2 tetramer
Method: single particle / : Sun Y, Ma SC, Chai JJ

EMDB-38685: 
Cryo-EM structure of tomato NRC2 filament
Method: helical / : Sun Y, Ma SC, Chai JJ

PDB-8xuq: 
Cryo-EM structure of tomato NRC2 tetramer
Method: single particle / : Sun Y, Ma SC, Chai JJ
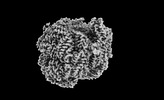
EMDB-41434: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA
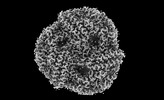
EMDB-41435: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA
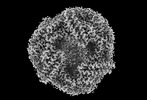
EMDB-41436: 
Central rod disk in D3 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA
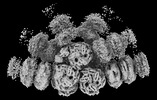
EMDB-41463: 
Synechocystis PCC 6803 Phycobilisome quenched by OCP, high resolution
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB-41475: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L
EMDB-41585: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to2: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to5: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search







 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
